Selective androgen receptor modulators activate the canonical prostate cancer androgen receptor program and repress cancer growth
- PMID: 33998604
- PMCID: PMC8121509
- DOI: 10.1172/JCI146777
Selective androgen receptor modulators activate the canonical prostate cancer androgen receptor program and repress cancer growth
Erratum in
-
Selective androgen receptor modulators activate the canonical prostate cancer androgen receptor program and repress cancer growth.J Clin Invest. 2021 Jun 15;131(12):e151719. doi: 10.1172/JCI151719. J Clin Invest. 2021. PMID: 34128479 Free PMC article. No abstract available.
Abstract
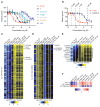
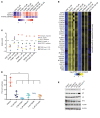
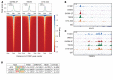
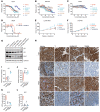
Prostate cancer (PC) is driven by androgen receptor (AR) activity, a master regulator of prostate development and homeostasis. Frontline therapies for metastatic PC deprive the AR of the activating ligands testosterone (T) and dihydrotestosterone (DHT) by limiting their biosynthesis or blocking AR binding. Notably, AR signaling is dichotomous, inducing growth at lower activity levels, while suppressing growth at higher levels. Recent clinical studies have exploited this effect by administration of supraphysiological concentrations of T, resulting in clinical responses and improvements in quality of life. However, the use of T as a therapeutic agent in oncology is limited by poor drug-like properties as well as rapid and variable metabolism. Here, we investigated the antitumor effects of selective AR modulators (SARMs), which are small-molecule nonsteroidal AR agonists developed to treat muscle wasting and cachexia. Several orally administered SARMs activated the AR program in PC models. AR cistromes regulated by steroidal androgens and SARMs were superimposable. Coregulatory proteins including HOXB13 and GRHL2 comprised AR complexes assembled by both androgens and SARMs. At bioavailable concentrations, SARMs repressed MYC oncoprotein expression and inhibited the growth of castration-sensitive and castration-resistant PC in vitro and in vivo. These results support further clinical investigation of SARMs for treating advanced PC.
Keywords: Oncology; Prostate cancer.
Conflict of interest statement
Figures



Comment in
-
Urological Oncology: Prostate Cancer.J Urol. 2021 Oct;206(4):1062-1064. doi: 10.1097/JU.0000000000002132. Epub 2021 Jul 20. J Urol. 2021. PMID: 34281353 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

