Rab40-Cullin5 complex regulates EPLIN and actin cytoskeleton dynamics during cell migration
- PMID: 33999101
- PMCID: PMC8129794
- DOI: 10.1083/jcb.202008060
Rab40-Cullin5 complex regulates EPLIN and actin cytoskeleton dynamics during cell migration
Abstract
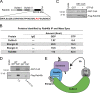
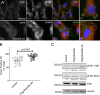
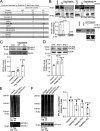
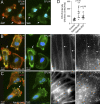
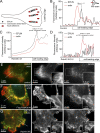
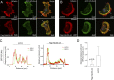
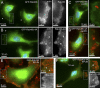
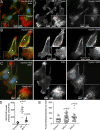
Rab40b is a SOCS box-containing protein that regulates the secretion of MMPs to facilitate extracellular matrix remodeling during cell migration. Here, we show that Rab40b interacts with Cullin5 via the Rab40b SOCS domain. We demonstrate that loss of Rab40b-Cullin5 binding decreases cell motility and invasive potential and show that defective cell migration and invasion stem from alteration to the actin cytoskeleton, leading to decreased invadopodia formation, decreased actin dynamics at the leading edge, and an increase in stress fibers. We also show that these stress fibers anchor at less dynamic, more stable focal adhesions. Mechanistically, changes in the cytoskeleton and focal adhesion dynamics are mediated in part by EPLIN, which we demonstrate to be a binding partner of Rab40b and a target for Rab40b-Cullin5-dependent localized ubiquitylation and degradation. Thus, we propose a model where Rab40b-Cullin5-dependent ubiquitylation regulates EPLIN localization to promote cell migration and invasion by altering focal adhesion and cytoskeletal dynamics.
© 2021 Linklater et al.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

