Genome-wide association mapping identifies yellow rust resistance loci in Ethiopian durum wheat germplasm
- PMID: 33999918
- PMCID: PMC8128278
- DOI: 10.1371/journal.pone.0243675
Genome-wide association mapping identifies yellow rust resistance loci in Ethiopian durum wheat germplasm
Abstract
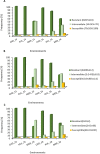
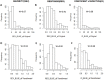
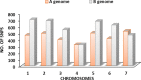
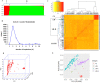
Durum wheat is an important cereal grown in Ethiopia, a country which is also its center for genetic diversity. Yellow (stripe) rust caused by Puccinia striiformis fsp tritici is one of the most devastating diseases threatening Ethiopian wheat production. To identify sources of genetic resistance and combat this pathogen, we conducted a genome wide association study of yellow rust resistance on 300 durum wheat accessions comprising 261 landraces and 39 cultivars. The accessions were evaluated for their field resistance using a modified Cobb scale at Meraro, Kulumsa and Chefe Donsa in the 2015 and 2016 main growing seasons. Analysis of the 35K Axiom Array genotyping data of the panel resulted in a total of 8,797 polymorphic SNPs of which 7,093 were used in subsequent analyses. Population structure analysis suggested two groups in which the cultivars clearly stood out separately from the landraces. Eleven SNPs significantly associated with yellow rust resistance were identified on four chromosomes (1A, 1B, 2B, and 5A) which defined at least five genomic loci. Six of the SNPs were consistently identified on chromosome 1B singly at each and combined overall environments which explained 62.6-64.0% of the phenotypic variation (R2). Resistant allele frequency ranged from 14.0-71.0%; Zooming in to the identified resistance loci revealed the presence of disease resistance related genes involved in the plant defense system such as the ABC transporter gene family, disease resistance protein RPM1 (NBS-LRR class), Receptor kinases and Protein kinases. This study has provided SNPs for tracking the loci associated with yellow rust resistance and a diversity panel which can be used for association study of other agriculturally important traits in durum wheat.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Chris G, World Durum Outlook. Available online: http://www.internationalpasta.org/ resources/ IPO%20BOARD%202013/2%20Chris%20Gillen.pdf.
-
- Vavilov NI. The origin, variation, immunity and breeding of cultivated plants. Soil Sci. 1951; 72:482.
-
- Vavilov NI, Dorofeev VF. Origin and Geography of Cultivated Plants. Cambridge: Cambridge University Press. 1992.
-
- Harlain JR. Ethiopia: a centre of diversity. Econ Bot. 1969; 23: 309–314.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

