Characterization of oral swab samples for diagnosis of pulmonary tuberculosis
- PMID: 33999938
- PMCID: PMC8128230
- DOI: 10.1371/journal.pone.0251422
Characterization of oral swab samples for diagnosis of pulmonary tuberculosis
Abstract
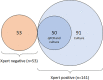
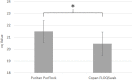
Oral swab analysis (OSA) has been shown to detect Mycobacterium tuberculosis (MTB) DNA in patients with pulmonary tuberculosis (TB). In previous analyses, qPCR testing of swab samples collected from tongue dorsa was up to 93% sensitive relative to sputum GeneXpert, when 2 swabs per patient were tested. The present study modified sample collection methods to increase sample biomass and characterized the viability of bacilli present in tongue swabs. A qPCR targeting conserved bacterial ribosomal rRNA gene (rDNA) sequences was used to quantify bacterial biomass in samples. There was no detectable reduction in total bacterial rDNA signal over the course of 10 rapidly repeated tongue samplings, indicating that swabs collect only a small portion of the biomass available for testing. Copan FLOQSwabs collected ~2-fold more biomass than Puritan PurFlock swabs, the best brand used previously (p = 0.006). FLOQSwabs were therefore evaluated in patients with possible TB in Uganda. A FLOQSwab was collected from each patient upon enrollment (Day 1) and, in a subset of sputum GeneXpert Ultra-positive patients, a second swab was collected on the following day (Day 2). Swabs were tested for MTB DNA by manual IS6110-targeted qPCR. Relative to sputum GeneXpert Ultra, single-swab sensitivity was 88% (44/50) on Day 1 and 94.4% (17/18) on Day 2. Specificity was 79.2% (42/53). Among an expanded sample of Ugandan patients, 62% (87/141) had colony-forming bacilli in their tongue dorsum swab samples. These findings will help guide further development of this promising TB screening method.
Conflict of interest statement
At the time of study design and evaluation SB, CMB, DB, and AS received salary support from the Global Good Fund. At the time of study design and evaluation GH, KPN, AML, CO, DM, and KJM were employed by Intellectual Ventures Laboratory. There are no patents, products in development or marketed products to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials. A number of the authors are currently affiliated with GH Labs and Roche Molecular Systems, but were not affiliated with them during the study, and these companies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Figures

References
-
- World Health Organization. Global Tuberculosis Report 2020 2020. 2020.
-
- UNITAID. Tuberculosis diagnostics technology and market landscape - 5th edition. World Health Organization. 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

