Light-dependent N-end rule-mediated disruption of protein function in Saccharomyces cerevisiae and Drosophila melanogaster
- PMID: 33999957
- PMCID: PMC8158876
- DOI: 10.1371/journal.pgen.1009544
Light-dependent N-end rule-mediated disruption of protein function in Saccharomyces cerevisiae and Drosophila melanogaster
Abstract
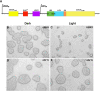
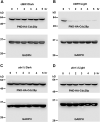
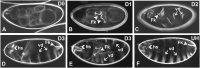
Here we describe the development and characterization of the photo-N-degron, a peptide tag that can be used in optogenetic studies of protein function in vivo. The photo-N-degron can be expressed as a genetic fusion to the amino termini of other proteins, where it undergoes a blue light-dependent conformational change that exposes a signal for the class of ubiquitin ligases, the N-recognins, which mediate the N-end rule mechanism of proteasomal degradation. We demonstrate that the photo-N-degron can be used to direct light-mediated degradation of proteins in Saccharomyces cerevisiae and Drosophila melanogaster with fine temporal control. In addition, we compare the effectiveness of the photo-N-degron with that of two other light-dependent degrons that have been developed in their abilities to mediate the loss of function of Cactus, a component of the dorsal-ventral patterning system in the Drosophila embryo. We find that like the photo-N-degron, the blue light-inducible degradation (B-LID) domain, a light-activated degron that must be placed at the carboxy terminus of targeted proteins, is also effective in eliciting light-dependent loss of Cactus function, as determined by embryonic dorsal-ventral patterning phenotypes. In contrast, another previously described photosensitive degron (psd), which also must be located at the carboxy terminus of associated proteins, has little effect on Cactus-dependent phenotypes in response to illumination of developing embryos. These and other observations indicate that care must be taken in the selection and application of light-dependent and other inducible degrons for use in studies of protein function in vivo, but importantly demonstrate that N- and C-terminal fusions to the photo-N-degron and the B-LID domain, respectively, support light-dependent degradation in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117: 1223–1237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

