Microbiota populations in supragingival plaque, subgingival plaque, and saliva habitats of adult dogs
- PMID: 34001282
- PMCID: PMC8130298
- DOI: 10.1186/s42523-021-00100-9
Microbiota populations in supragingival plaque, subgingival plaque, and saliva habitats of adult dogs
Erratum in
-
Correction to: Microbiota populations in supragingival plaque, subgingival plaque, and saliva habitats of adult dogs.Anim Microbiome. 2021 Jun 3;3(1):40. doi: 10.1186/s42523-021-00102-7. Anim Microbiome. 2021. PMID: 34082838 Free PMC article. No abstract available.
Abstract
Background: Oral diseases are common in dogs, with microbiota playing a prominent role in the disease process. Oral cavity habitats harbor unique microbiota populations that have relevance to health and disease. Despite their importance, the canine oral cavity microbial habitats have been poorly studied. The objectives of this study were to (1) characterize the oral microbiota of different habitats of dogs and (2) correlate oral health scores with bacterial taxa and identify what sites may be good options for understanding the role of microbiota in oral diseases. We used next-generation sequencing to characterize the salivary (SAL), subgingival (SUB), and supragingival (SUP) microbial habitats of 26 healthy adult female Beagle dogs (4.0 ± 1.2 year old) and identify taxa associated with periodontal disease indices.
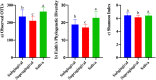
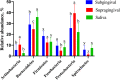
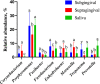
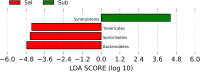
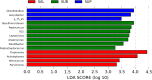
Results: Bacterial species richness was highest for SAL, moderate for SUB, and lowest for SUP samples (p < 0.001). Unweighted and weighted principal coordinates plots showed clustering by habitat, with SAL and SUP samples being the most different from one another. Bacteroidetes, Proteobacteria, Firmicutes, Fusobacteria, Actinobacteria, and Spirochaetes were the predominant phyla in all habitats. Paludibacter, Filifactor, Peptostreptococcus, Fusibacter, Anaerovorax, Fusobacterium, Leptotrichia, Desulfomicrobium, and TG5 were enriched in SUB samples, while Actinomyces, Corynebacterium, Leucobacter, Euzebya, Capnocytophaga, Bergeyella, Lautropia, Lampropedia, Desulfobulbus, Enhydrobacter, and Moraxella were enriched in SUP samples. Prevotella, SHD-231, Helcococcus, Treponema, and Acholeplasma were enriched in SAL samples. p-75-a5, Arcobacter, and Pasteurella were diminished in SUB samples. Porphyromonas, Peptococcus, Parvimonas, and Campylobacter were diminished in SUP samples, while Tannerella, Proteocalla, Schwartzia, and Neisseria were diminished in SAL samples. Actinomyces, Corynebacterium, Capnocytophaga, Leptotrichia, and Neisseria were associated with higher oral health scores (worsened health) in plaque samples.
Conclusions: Our results demonstrate the differences that exist among canine salivary, subgingival plaque and supragingival plaque habitats. Salivary samples do not require sedation and are easy to collect, but do not accurately represent the plaque populations that are most important to oral disease. Plaque Actinomyces, Corynebacterium, Capnocytophaga, Leptotrichia, and Neisseria were associated with higher (worse) oral health scores. Future studies analyzing samples from progressive disease stages are needed to validate these results and understand the role of bacteria in periodontal disease development.
Keywords: 16S rRNA gene; Canine; Oral microbiome; Periodontal disease.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Oral microbiota populations of adult dogs consuming wet or dry foods.J Anim Sci. 2022 Aug 1;100(8):skac200. doi: 10.1093/jas/skac200. J Anim Sci. 2022. PMID: 35641105 Free PMC article.
-
Dental chews positively shift the oral microbiota of adult dogs.J Anim Sci. 2021 Jul 1;99(7):skab100. doi: 10.1093/jas/skab100. J Anim Sci. 2021. PMID: 33780530 Free PMC article.
-
Effects of a novel dental chew on oral health outcomes, halitosis, and microbiota of adult dogs.J Anim Sci. 2024 Jan 3;102:skae071. doi: 10.1093/jas/skae071. J Anim Sci. 2024. PMID: 38477668 Free PMC article.
-
An overview of the microbiota of the oral cavity of humans and non-human primates with periodontal disease: Current issues and perspectives.Arch Oral Biol. 2025 Apr;172:106121. doi: 10.1016/j.archoralbio.2024.106121. Epub 2024 Nov 10. Arch Oral Biol. 2025. PMID: 39808970 Review.
-
Dental calculus: recent insights into occurrence, formation, prevention, removal and oral health effects of supragingival and subgingival deposits.Eur J Oral Sci. 1997 Oct;105(5 Pt 2):508-22. doi: 10.1111/j.1600-0722.1997.tb00238.x. Eur J Oral Sci. 1997. PMID: 9395117 Review.
Cited by
-
The Relationship of Tumor Microbiome and Oral Bacteria and Intestinal Dysbiosis in Canine Mammary Tumor.Int J Mol Sci. 2022 Sep 18;23(18):10928. doi: 10.3390/ijms231810928. Int J Mol Sci. 2022. PMID: 36142841 Free PMC article.
-
From wolves to humans: oral microbiome resistance to transfer across mammalian hosts.mBio. 2024 Mar 13;15(3):e0334223. doi: 10.1128/mbio.03342-23. Epub 2024 Feb 1. mBio. 2024. PMID: 38299854 Free PMC article.
-
Species-level characterization of saliva and dental plaque microbiota reveals putative bacterial and functional biomarkers of periodontal diseases in dogs.FEMS Microbiol Ecol. 2024 May 14;100(6):fiae082. doi: 10.1093/femsec/fiae082. FEMS Microbiol Ecol. 2024. PMID: 38782729 Free PMC article.
-
Oral microbiota populations of adult dogs consuming wet or dry foods.J Anim Sci. 2022 Aug 1;100(8):skac200. doi: 10.1093/jas/skac200. J Anim Sci. 2022. PMID: 35641105 Free PMC article.
-
Influence of Gallic Acid-Containing Mouth Spray on Dental Health and Oral Microbiota of Healthy Dogs: A Pilot Study.Vet Sci. 2023 Jun 30;10(7):424. doi: 10.3390/vetsci10070424. Vet Sci. 2023. PMID: 37505829 Free PMC article.
References
-
- Butković V, Šehič M, Stanin D, Šimpraga M, Capak D, Kos J. Dental diseases in dogs: a retrospective study of radiological data. Acta Vet Brno. 2001;70:203–8. doi: 10.2754/avb200170020203. - DOI
-
- Kyllar M, Witter K. Prevalence of dental disorders in pet dogs. Vet Med (Praha) 2005;50:496–505. doi: 10.17221/5654-VETMED. - DOI
-
- Hamp S-E, Olsson S-E, Farsø-Madsen K, Viklands P, Fornell J. A macroscopic and radiological investigation of dental diseases of the dog. Vet Radiol. 1984;25:86–92. doi: 10.1111/j.1740-8261.1984.tb01916.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
