Piperidine-4-Carboxamides Target DNA Gyrase in Mycobacterium abscessus
- PMID: 34001512
- PMCID: PMC8284461
- DOI: 10.1128/AAC.00676-21
Piperidine-4-Carboxamides Target DNA Gyrase in Mycobacterium abscessus
Abstract
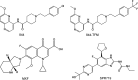
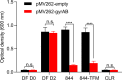
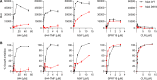
New, more-effective drugs for the treatment of lung disease caused by nontuberculous mycobacteria (NTM) are needed. Among NTM opportunistic pathogens, Mycobacterium abscessus is the most difficult to cure and intrinsically multidrug resistant. In a whole-cell screen of a compound collection active against Mycobacterium tuberculosis, we previously identified the piperidine-4-carboxamide (P4C) MMV688844 (844) as a hit against M. abscessus. Here, we identified a more potent analog of 844 and showed that both the parent and improved analog retain activity against strains representing all three subspecies of the M. abscessus complex. Furthermore, P4Cs showed bactericidal and antibiofilm activity. Spontaneous resistance against the P4Cs emerged at a frequency of 10-8/CFU and mapped to gyrA and gyrB encoding the subunits of DNA gyrase. Biochemical studies with recombinant M. abscessus DNA gyrase showed that P4Cs inhibit the wild-type enzyme but not the P4C-resistant mutant. P4C-resistant strains showed limited cross-resistance to the fluoroquinolone moxifloxacin, which is in clinical use for the treatment of macrolide-resistant M. abscessus disease, and no cross-resistance to the benzimidazole SPR719, a novel DNA gyrase inhibitor in clinical development for the treatment of mycobacterial diseases. Analyses of P4Cs in recA promoter-based DNA damage reporter strains showed induction of recA promoter activity in the wild type but not in the P4C-resistant mutant background. This indicates that P4Cs, similar to fluoroquinolones, cause DNA gyrase-mediated DNA damage. Together, our results show that P4Cs present a novel class of mycobacterial DNA gyrase inhibitors with attractive antimicrobial activities against the M. abscessus complex.
Keywords: DNA gyrase; MMV688844; Mycobacterium abscessus; NTM; nontuberculous mycobacteria.
Figures



Similar articles
-
Activity of Tricyclic Pyrrolopyrimidine Gyrase B Inhibitor against Mycobacterium abscessus.Antimicrob Agents Chemother. 2022 Sep 20;66(9):e0066922. doi: 10.1128/aac.00669-22. Epub 2022 Aug 25. Antimicrob Agents Chemother. 2022. PMID: 36005813 Free PMC article.
-
In Vitro Resistance against DNA Gyrase Inhibitor SPR719 in Mycobacterium avium and Mycobacterium abscessus.Microbiol Spectr. 2022 Feb 23;10(1):e0132121. doi: 10.1128/spectrum.01321-21. Epub 2022 Jan 12. Microbiol Spectr. 2022. PMID: 35019671 Free PMC article.
-
A Mycobacterium tuberculosis NBTI DNA Gyrase Inhibitor Is Active against Mycobacterium abscessus.Antimicrob Agents Chemother. 2021 Nov 17;65(12):e0151421. doi: 10.1128/AAC.01514-21. Epub 2021 Oct 4. Antimicrob Agents Chemother. 2021. PMID: 34606340 Free PMC article.
-
Mycobacterium abscessus: insights from a bioinformatic perspective.Crit Rev Microbiol. 2023 Aug;49(4):499-514. doi: 10.1080/1040841X.2022.2082268. Epub 2022 Jun 13. Crit Rev Microbiol. 2023. PMID: 35696783 Review.
-
The treatment of rapidly growing mycobacterial infections.Clin Chest Med. 2015 Mar;36(1):67-78. doi: 10.1016/j.ccm.2014.10.004. Epub 2014 Nov 5. Clin Chest Med. 2015. PMID: 25676520 Review.
Cited by
-
Activity of Tricyclic Pyrrolopyrimidine Gyrase B Inhibitor against Mycobacterium abscessus.Antimicrob Agents Chemother. 2022 Sep 20;66(9):e0066922. doi: 10.1128/aac.00669-22. Epub 2022 Aug 25. Antimicrob Agents Chemother. 2022. PMID: 36005813 Free PMC article.
-
Durlobactam to boost the clinical utility of standard of care β-lactams against Mycobacterium abscessus lung disease.Antimicrob Agents Chemother. 2025 Jan 31;69(1):e0104624. doi: 10.1128/aac.01046-24. Epub 2024 Nov 20. Antimicrob Agents Chemother. 2025. PMID: 39565116 Free PMC article.
-
Why Matter Matters: Fast-Tracking Mycobacterium abscessus Drug Discovery.Molecules. 2022 Oct 17;27(20):6948. doi: 10.3390/molecules27206948. Molecules. 2022. PMID: 36296540 Free PMC article. Review.
-
Cyclohexyl-griselimycin Is Active against Mycobacterium abscessus in Mice.Antimicrob Agents Chemother. 2022 Jan 18;66(1):e0140021. doi: 10.1128/AAC.01400-21. Epub 2021 Nov 1. Antimicrob Agents Chemother. 2022. PMID: 34723632 Free PMC article.
-
Toward a Bactericidal Oral Drug Combination for the Treatment of Mycobacterium abscessus Lung Disease.ACS Infect Dis. 2025 Apr 11;11(4):929-939. doi: 10.1021/acsinfecdis.4c00948. Epub 2025 Apr 1. ACS Infect Dis. 2025. PMID: 40168319 Free PMC article.
References
-
- Tortoli E. 2019. The taxonomy of the genus mycobacterium, p 1–10. In Velayati AA, Farnia P (ed), Nontuberculous mycobacteria (NTM). Academic Press, New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

