mascRNA and its parent lncRNA MALAT1 promote proliferation and metastasis of hepatocellular carcinoma cells by activating ERK/MAPK signaling pathway
- PMID: 34001866
- PMCID: PMC8128908
- DOI: 10.1038/s41420-021-00497-x
mascRNA and its parent lncRNA MALAT1 promote proliferation and metastasis of hepatocellular carcinoma cells by activating ERK/MAPK signaling pathway
Abstract
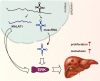
MALAT1-associated small cytoplasmic RNA (mascRNA) is a cytoplasmic tRNA-like small RNA derived from nucleus-located long noncoding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1). While MALAT1 was extensively studied and was found to function in multiple cellular processes, including tumorigenesis and tumor progression, the role of mascRNA was largely unknown. Here we show that mascRNA is upregulated in multiple cancer cell lines and hepatocellular carcinoma (HCC) clinical samples. Using HCC cells as model, we found that mascRNA and its parent lncRNA MALAT1 can both promote cell proliferation, migration, and invasion in vitro. Correspondingly, both of them can enhance the tumor growth in mice subcutaneous tumor model and can promote metastasis by tail intravenous injection of HCC cells. Furthermore, we revealed that mascRNA and MALAT1 can both activate ERK/MAPK signaling pathway, which regulates metastasis-related genes and may contribute to the aggressive phenotype of HCC cells. Our results indicate a coordination in function and mechanism of mascRNA and MALAT1 during development and progress of HCC, and provide a paradigm for deciphering tRNA-like structures and their parent transcripts in mammalian cells.
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
The tRNA-like small noncoding RNA mascRNA promotes global protein translation.EMBO Rep. 2020 Dec 3;21(12):e49684. doi: 10.15252/embr.201949684. Epub 2020 Oct 19. EMBO Rep. 2020. PMID: 33073493 Free PMC article.
-
miR-124-3p availability is antagonized by LncRNA-MALAT1 for Slug-induced tumor metastasis in hepatocellular carcinoma.Cancer Med. 2019 Oct;8(14):6358-6369. doi: 10.1002/cam4.2482. Epub 2019 Aug 29. Cancer Med. 2019. PMID: 31466138 Free PMC article.
-
Structural basis of MALAT1 RNA maturation and mascRNA biogenesis.Nat Struct Mol Biol. 2024 Nov;31(11):1655-1668. doi: 10.1038/s41594-024-01340-4. Epub 2024 Jul 2. Nat Struct Mol Biol. 2024. PMID: 38956168
-
Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma.Biomed Pharmacother. 2018 Jun;102:653-669. doi: 10.1016/j.biopha.2018.03.105. Epub 2018 Apr 5. Biomed Pharmacother. 2018. PMID: 29604585
-
Long noncoding RNAs: Novel insights into hepatocelluar carcinoma.Cancer Lett. 2014 Mar 1;344(1):20-27. doi: 10.1016/j.canlet.2013.10.021. Epub 2013 Oct 30. Cancer Lett. 2014. PMID: 24183851 Review.
Cited by
-
Diversity of Dysregulated Long Non-Coding RNAs in HBV-Related Hepatocellular Carcinoma.Front Immunol. 2022 Jan 28;13:834650. doi: 10.3389/fimmu.2022.834650. eCollection 2022. Front Immunol. 2022. PMID: 35154157 Free PMC article. Review.
-
The Diagnosis and Prognosis Value of Exosomal MascRNA in Patients with Acute Coronary Syndrome.Int J Gen Med. 2025 Aug 12;18:4425-4436. doi: 10.2147/IJGM.S538127. eCollection 2025. Int J Gen Med. 2025. PMID: 40822895 Free PMC article.
-
Ginsenoside Rh3 Inhibits Lung Cancer Metastasis by Targeting Extracellular Signal-Regulated Kinase: A Network Pharmacology Study.Pharmaceuticals (Basel). 2022 Jun 17;15(6):758. doi: 10.3390/ph15060758. Pharmaceuticals (Basel). 2022. PMID: 35745677 Free PMC article.
-
Absence of Scaffold Protein Tks4 Disrupts Several Signaling Pathways in Colon Cancer Cells.Int J Mol Sci. 2023 Jan 9;24(2):1310. doi: 10.3390/ijms24021310. Int J Mol Sci. 2023. PMID: 36674824 Free PMC article.
-
LINC00478-derived novel cytoplasmic lncRNA LacRNA stabilizes PHB2 and suppresses breast cancer metastasis via repressing MYC targets.J Transl Med. 2023 Feb 13;21(1):120. doi: 10.1186/s12967-023-03967-1. J Transl Med. 2023. PMID: 36782197 Free PMC article.
References
Grants and funding
- 2021A1515010477/Natural Science Foundation of Guangdong Province (Guangdong Natural Science Foundation)
- 2018M643347/China Postdoctoral Science Foundation
- 81402426/National Natural Science Foundation of China (National Science Foundation of China)
- 81870449/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

