Connectivity characterization of the mouse basolateral amygdalar complex
- PMID: 34001873
- PMCID: PMC8129205
- DOI: 10.1038/s41467-021-22915-5
Connectivity characterization of the mouse basolateral amygdalar complex
Abstract
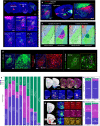
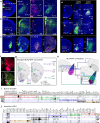
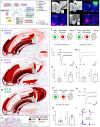
The basolateral amygdalar complex (BLA) is implicated in behaviors ranging from fear acquisition to addiction. Optogenetic methods have enabled the association of circuit-specific functions to uniquely connected BLA cell types. Thus, a systematic and detailed connectivity profile of BLA projection neurons to inform granular, cell type-specific interrogations is warranted. Here, we apply machine-learning based computational and informatics analysis techniques to the results of circuit-tracing experiments to create a foundational, comprehensive BLA connectivity map. The analyses identify three distinct domains within the anterior BLA (BLAa) that house target-specific projection neurons with distinguishable morphological features. We identify brain-wide targets of projection neurons in the three BLAa domains, as well as in the posterior BLA, ventral BLA, posterior basomedial, and lateral amygdalar nuclei. Inputs to each nucleus also are identified via retrograde tracing. The data suggests that connectionally unique, domain-specific BLAa neurons are associated with distinct behavior networks.
Conflict of interest statement
The authors declare no competing interests.
Figures






Comment in
-
Brain Networks of Connectionally Unique Basolateral Amygdala Cell Types.Neurosci Insights. 2022 Feb 26;17:26331055221080175. doi: 10.1177/26331055221080175. eCollection 2022. Neurosci Insights. 2022. PMID: 35252870 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

