Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma
- PMID: 34001881
- PMCID: PMC8128874
- DOI: 10.1038/s41467-021-22872-z
Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma
Erratum in
-
Author Correction: Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma.Nat Commun. 2024 Oct 7;15(1):8668. doi: 10.1038/s41467-024-52013-1. Nat Commun. 2024. PMID: 39375332 Free PMC article. No abstract available.
Abstract
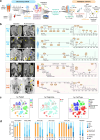
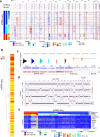
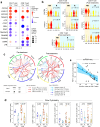
The mechanisms driving therapeutic resistance and poor outcomes of mantle cell lymphoma (MCL) are incompletely understood. We characterize the cellular and molecular heterogeneity within and across patients and delineate the dynamic evolution of tumor and immune cell compartments at single cell resolution in longitudinal specimens from ibrutinib-sensitive patients and non-responders. Temporal activation of multiple cancer hallmark pathways and acquisition of 17q are observed in a refractory MCL. Multi-platform validation is performed at genomic and cellular levels in PDX models and larger patient cohorts. We demonstrate that due to 17q gain, BIRC5/survivin expression is upregulated in resistant MCL tumor cells and targeting BIRC5 results in marked tumor inhibition in preclinical models. In addition, we discover notable differences in the tumor microenvironment including progressive dampening of CD8+ T cells and aberrant cell-to-cell communication networks in refractory MCLs. This study reveals diverse and dynamic tumor and immune programs underlying therapy resistance in MCL.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Argatoff, L. H., Connors, J. M., Klasa, R. J., Horsman, D. E. & Gascoyne, R. D. Mantle cell lymphoma: a clinicopathologic study of 80 cases. Blood89, 2067–2078 (1997). - PubMed
-
- Gujral, S. et al. A clinicopathologic study of mantle cell lymphoma in a single center study in India. Indian J. Pathol. Microbiol.51, 315–322 (2008). - PubMed
-
- Hankin, R. C. & Hunter, S. V. Mantle cell lymphoma. Arch. Pathol. Lab. Med.123, 1182–1188 (1999). - PubMed
-
- Kauh, J., Baidas, S. M., Ozdemirli, M. & Cheson, B. D. Mantle cell lymphoma: clinicopathologic features and treatments. Oncology17, 879–891 (2003). 896; discussion 896–878. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

