Genetic fate-mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury
- PMID: 34001904
- PMCID: PMC8129080
- DOI: 10.1038/s41467-021-23197-7
Genetic fate-mapping reveals surface accumulation but not deep organ invasion of pleural and peritoneal cavity macrophages following injury
Abstract
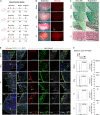
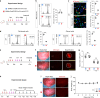
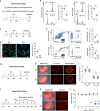
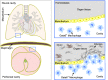
During injury, monocytes are recruited from the circulation to inflamed tissues and differentiate locally into mature macrophages, with prior reports showing that cavity macrophages of the peritoneum and pericardium invade deeply into the respective organs to promote repair. Here we report a dual recombinase-mediated genetic system designed to trace cavity macrophages in vivo by intersectional detection of two characteristic markers. Lineage tracing with this method shows accumulation of cavity macrophages during lung and liver injury on the surface of visceral organs without penetration into the parenchyma. Additional data suggest that these peritoneal or pleural cavity macrophages do not contribute to tissue repair and regeneration. Our in vivo genetic targeting approach thus provides a reliable method to identify and characterize cavity macrophages during their development and in tissue repair and regeneration, and distinguishes these cells from other lineages.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

