Characterization of the human TARDBP gene promoter
- PMID: 34002018
- PMCID: PMC8129075
- DOI: 10.1038/s41598-021-89973-z
Characterization of the human TARDBP gene promoter
Abstract
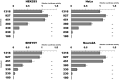
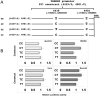
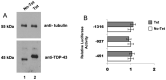
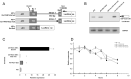
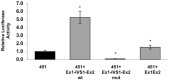
The expression of TDP-43, the main component of neuronal intracellular inclusions across a broad spectrum of ALS and FTD disorders, is developmentally regulated and studies in vivo have shown that TDP-43 overexpression can be toxic, even before observation of pathological aggregates. Starting from these observations, the regulation of its expression at transcriptional level might represent a further key element for the pathogenesis of neurodegenerative diseases. Therefore, we have characterized the human TARDBP promoter, in order to study the transcriptional mechanisms of expression. Mapping of cis-acting elements by luciferase assays in different cell outlined that the activity of the promoter seems to be higher in SH-SY5Y, Neuro2A, and HeLa than in HEK293. In addition, we tested effects of two SNPs found in the promoter region of ALS patients and observed no significant effect on transcription levels in all tested cell lines. Lastly, while TDP-43 overexpression did not affect significantly the activity of its promoter (suggesting that TDP-43 does not influence its own transcription), the presence of the 5'UTR sequence and of intron-1 splicing seem to impact positively on TDP-43 expression without affecting transcript stability. In conclusion, we have identified the region spanning nucleotides 451-230 upstream from the transcription start site as the minimal region with a significant transcription activity. These results lay an important foundation for exploring the regulation of the TARDBP gene transcription by exogenous and endogenous stimuli and the implication of transcriptional mechanisms in the pathogenesis of TDP-43 proteinopathies.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Characterization of the upstream and intron promoters of the gene encoding TAR DNA-binding protein.Sci Rep. 2021 Apr 22;11(1):8720. doi: 10.1038/s41598-021-88015-y. Sci Rep. 2021. PMID: 33888768 Free PMC article.
-
TARDBP mutation analysis in TDP-43 proteinopathies and deciphering the toxicity of mutant TDP-43.J Alzheimers Dis. 2013;33 Suppl 1(Suppl 1):S35-45. doi: 10.3233/JAD-2012-129036. J Alzheimers Dis. 2013. PMID: 22751173 Free PMC article. Review.
-
A novel Drosophila model of TDP-43 proteinopathies: N-terminal sequences combined with the Q/N domain induce protein functional loss and locomotion defects.Dis Model Mech. 2016 Jun 1;9(6):659-69. doi: 10.1242/dmm.023382. Epub 2016 Apr 21. Dis Model Mech. 2016. PMID: 27101846 Free PMC article.
-
Promoter activity of SARS coronavirus 5' UTR sequence in eukaryotic cells.Sichuan Da Xue Xue Bao Yi Xue Ban. 2006 Jan;37(1):5-9. Sichuan Da Xue Xue Bao Yi Xue Ban. 2006. PMID: 16468630
-
[TDP-43 proteinopathies, toward understanding of the molecular pathogenesis].Rinsho Shinkeigaku. 2009 Nov;49(11):783-5. doi: 10.5692/clinicalneurol.49.783. Rinsho Shinkeigaku. 2009. PMID: 20030209 Review. Japanese.
Cited by
-
TDP-43 Epigenetic Facets and Their Neurodegenerative Implications.Int J Mol Sci. 2023 Sep 7;24(18):13807. doi: 10.3390/ijms241813807. Int J Mol Sci. 2023. PMID: 37762112 Free PMC article. Review.
-
TARDBP Inhibits Porcine Epidemic Diarrhea Virus Replication through Degrading Viral Nucleocapsid Protein and Activating Type I Interferon Signaling.J Virol. 2022 May 25;96(10):e0007022. doi: 10.1128/jvi.00070-22. Epub 2022 May 2. J Virol. 2022. PMID: 35499322 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

