Functionally distinct POMC-expressing neuron subpopulations in hypothalamus revealed by intersectional targeting
- PMID: 34002087
- PMCID: PMC8249241
- DOI: 10.1038/s41593-021-00854-0
Functionally distinct POMC-expressing neuron subpopulations in hypothalamus revealed by intersectional targeting
Abstract
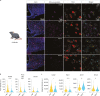
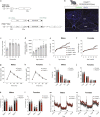
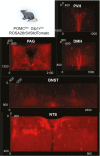
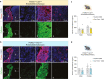
Pro-opiomelanocortin (POMC)-expressing neurons in the arcuate nucleus of the hypothalamus represent key regulators of metabolic homeostasis. Electrophysiological and single-cell sequencing experiments have revealed a remarkable degree of heterogeneity of these neurons. However, the exact molecular basis and functional consequences of this heterogeneity have not yet been addressed. Here, we have developed new mouse models in which intersectional Cre/Dre-dependent recombination allowed for successful labeling, translational profiling and functional characterization of distinct POMC neurons expressing the leptin receptor (Lepr) and glucagon like peptide 1 receptor (Glp1r). Our experiments reveal that POMCLepr+ and POMCGlp1r+ neurons represent largely nonoverlapping subpopulations with distinct basic electrophysiological properties. They exhibit a specific anatomical distribution within the arcuate nucleus and differentially express receptors for energy-state communicating hormones and neurotransmitters. Finally, we identify a differential ability of these subpopulations to suppress feeding. Collectively, we reveal a notably distinct functional microarchitecture of critical metabolism-regulatory neurons.
Conflict of interest statement
The authors declare no competing interests.
Figures
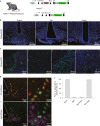
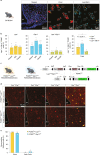
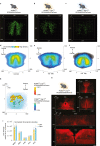
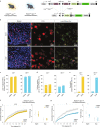









References
-
- Cowley MA, et al. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature. 2001;411:480–484. - PubMed
-
- Boston BA, Blaydon KM, Varnerin J, Cone RD. Independent and additive effects of central POMC and leptin pathways on murine obesity. Science. 1997;278:1641–1644. - PubMed
-
- Balthasar N, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. - PubMed
-
- Barsh G, et al. Neuroendocrine regulation by the Agouti/Agrp-melanocortin system. Endocr. Res. 2000;26:571. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

