An atlas of mitochondrial DNA genotype-phenotype associations in the UK Biobank
- PMID: 34002094
- PMCID: PMC7611844
- DOI: 10.1038/s41588-021-00868-1
An atlas of mitochondrial DNA genotype-phenotype associations in the UK Biobank
Abstract
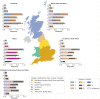
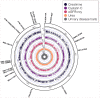
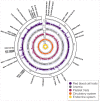
Mitochondrial DNA (mtDNA) variation in common diseases has been underexplored, partly due to a lack of genotype calling and quality-control procedures. Developing an at-scale workflow for mtDNA variant analyses, we show correlations between nuclear and mitochondrial genomic structures within subpopulations of Great Britain and establish a UK Biobank reference atlas of mtDNA-phenotype associations. A total of 260 mtDNA-phenotype associations were new (P < 1 × 10-5), including rs2853822 /m.8655 C>T (MT-ATP6) with type 2 diabetes, rs878966690 /m.13117 A>G (MT-ND5) with multiple sclerosis, 6 mtDNA associations with adult height, 24 mtDNA associations with 2 liver biomarkers and 16 mtDNA associations with parameters of renal function. Rare-variant gene-based tests implicated complex I genes modulating mean corpuscular volume and mean corpuscular hemoglobin. Seven traits had both rare and common mtDNA associations, where rare variants tended to have larger effects than common variants. Our work illustrates the value of studying mtDNA variants in common complex diseases and lays foundations for future large-scale mtDNA association studies.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
JMMH and EYD become full time employees of Novo Nordisk during the drafting of the manuscript. The remaining authors declare no conflicts of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

