Comparative Genomics Reveals Factors Associated with Phenotypic Expression of Wolbachia
- PMID: 34003269
- PMCID: PMC8290115
- DOI: 10.1093/gbe/evab111
Comparative Genomics Reveals Factors Associated with Phenotypic Expression of Wolbachia
Abstract
Wolbachia is a widespread, vertically transmitted bacterial endosymbiont known for manipulating arthropod reproduction. Its most common form of reproductive manipulation is cytoplasmic incompatibility (CI), observed when a modification in the male sperm leads to embryonic lethality unless a compatible rescue factor is present in the female egg. CI attracts scientific attention due to its implications for host speciation and in the use of Wolbachia for controlling vector-borne diseases. However, our understanding of CI is complicated by the complexity of the phenotype, whose expression depends on both symbiont and host factors. In the present study, we perform a comparative analysis of nine complete Wolbachia genomes with known CI properties in the same genetic host background, Drosophila simulans STC. We describe genetic differences between closely related strains and uncover evidence that phages and other mobile elements contribute to the rapid evolution of both genomes and phenotypes of Wolbachia. Additionally, we identify both known and novel genes associated with the modification and rescue functions of CI. We combine our observations with published phenotypic information and discuss how variability in cif genes, novel CI-associated genes, and Wolbachia titer might contribute to poorly understood aspects of CI such as strength and bidirectional incompatibility. We speculate that high titer CI strains could be better at invading new hosts already infected with a CI Wolbachia, due to a higher rescue potential, and suggest that titer might thus be a relevant parameter to consider for future strategies using CI Wolbachia in biological control.
Keywords: Drosophila; Wolbachia; comparative genomics; cytoplasmic incompatibility; genomics; symbiosis.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
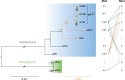
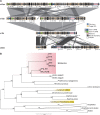

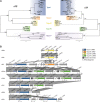
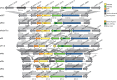
References
-
- ΘAbascal F, Valencia A.. 2003. Automatic annotation of protein function based on family identification. Proteins 53(3):683–692. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

