Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19
- PMID: 34003804
- PMCID: PMC8245175
- DOI: 10.1172/JCI148517
Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19
Abstract
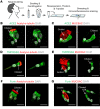
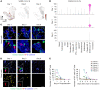
The upper respiratory tract is compromised in the early period of COVID-19, but SARS-CoV-2 tropism at the cellular level is not fully defined. Unlike recent single-cell RNA-Seq analyses indicating uniformly low mRNA expression of SARS-CoV-2 entry-related host molecules in all nasal epithelial cells, we show that the protein levels are relatively high and that their localizations are restricted to the apical side of multiciliated epithelial cells. In addition, we provide evidence in patients with COVID-19 that SARS-CoV-2 is massively detected and replicated within the multiciliated cells. We observed these findings during the early stage of COVID-19, when infected ciliated cells were rapidly replaced by differentiating precursor cells. Moreover, our analyses revealed that SARS-CoV-2 cellular tropism was restricted to the nasal ciliated versus oral squamous epithelium. These results imply that targeting ciliated cells of the nasal epithelium during the early stage of COVID-19 could be an ideal strategy to prevent SARS-CoV-2 propagation.
Keywords: COVID-19; Infectious disease; Molecular pathology.
Conflict of interest statement
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

