Real-time single-cell characterization of the eukaryotic transcription cycle reveals correlations between RNA initiation, elongation, and cleavage
- PMID: 34003867
- PMCID: PMC8162642
- DOI: 10.1371/journal.pcbi.1008999
Real-time single-cell characterization of the eukaryotic transcription cycle reveals correlations between RNA initiation, elongation, and cleavage
Abstract
The eukaryotic transcription cycle consists of three main steps: initiation, elongation, and cleavage of the nascent RNA transcript. Although each of these steps can be regulated as well as coupled with each other, their in vivo dissection has remained challenging because available experimental readouts lack sufficient spatiotemporal resolution to separate the contributions from each of these steps. Here, we describe a novel application of Bayesian inference techniques to simultaneously infer the effective parameters of the transcription cycle in real time and at the single-cell level using a two-color MS2/PP7 reporter gene and the developing fruit fly embryo as a case study. Our method enables detailed investigations into cell-to-cell variability in transcription-cycle parameters as well as single-cell correlations between these parameters. These measurements, combined with theoretical modeling, suggest a substantial variability in the elongation rate of individual RNA polymerase molecules. We further illustrate the power of this technique by uncovering a novel mechanistic connection between RNA polymerase density and nascent RNA cleavage efficiency. Thus, our approach makes it possible to shed light on the regulatory mechanisms in play during each step of the transcription cycle in individual, living cells at high spatiotemporal resolution.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

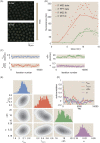

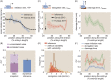

Similar articles
-
The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling.Biochim Biophys Acta. 2012 May;1819(5):391-400. doi: 10.1016/j.bbagrm.2012.01.010. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22306664 Review.
-
The RNA polymerase II elongation complex. Factor-dependent transcription elongation involves nascent RNA cleavage.J Biol Chem. 1992 Aug 5;267(22):15516-22. J Biol Chem. 1992. PMID: 1379232 Free PMC article.
-
Transcription factors TFIIF, ELL, and Elongin negatively regulate SII-induced nascent transcript cleavage by non-arrested RNA polymerase II elongation intermediates.J Biol Chem. 2001 Jun 22;276(25):23109-14. doi: 10.1074/jbc.M101445200. Epub 2001 Mar 19. J Biol Chem. 2001. PMID: 11259417
-
Measuring transcription dynamics in living cells using fluctuation analysis.Methods Mol Biol. 2013;1042:47-60. doi: 10.1007/978-1-62703-526-2_4. Methods Mol Biol. 2013. PMID: 23979999 Free PMC article.
-
Promoting elongation with transcript cleavage stimulatory factors.Biochim Biophys Acta. 2002 Sep 13;1577(2):287-307. doi: 10.1016/s0167-4781(02)00459-1. Biochim Biophys Acta. 2002. PMID: 12213659 Review.
Cited by
-
Gene expression model inference from snapshot RNA data using Bayesian non-parametrics.Nat Comput Sci. 2023 Feb;3(2):174-183. doi: 10.1038/s43588-022-00392-0. Epub 2023 Jan 19. Nat Comput Sci. 2023. PMID: 38125199 Free PMC article.
-
Chromatin regulates alternative polyadenylation via the RNA polymerase II elongation rate.Proc Natl Acad Sci U S A. 2024 May 21;121(21):e2405827121. doi: 10.1073/pnas.2405827121. Epub 2024 May 15. Proc Natl Acad Sci U S A. 2024. PMID: 38748572 Free PMC article.
-
Gene activity fully predicts transcriptional bursting dynamics.ArXiv [Preprint]. 2024 Jun 28:arXiv:2304.08770v3. ArXiv. 2024. PMID: 37131882 Free PMC article. Preprint.
-
Notch-dependent and -independent transcription are modulated by tissue movements at gastrulation.Elife. 2022 May 18;11:e73656. doi: 10.7554/eLife.73656. Elife. 2022. PMID: 35583918 Free PMC article.
-
Systematic inference identifies a major source of heterogeneity in cell signaling dynamics: The rate-limiting step number.Sci Adv. 2022 Mar 18;8(11):eabl4598. doi: 10.1126/sciadv.abl4598. Epub 2022 Mar 18. Sci Adv. 2022. PMID: 35302852 Free PMC article.
References
-
- Alberts B. Molecular biology of the cell. Sixth edition. ed. New York, NY: Garland Science, Taylor and Francis Group; 2015.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

