EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in Arabidopsis
- PMID: 34003929
- PMCID: PMC8408475
- DOI: 10.1093/plcell/koab139
EAR domain-containing transcription factors trigger PRC2-mediated chromatin marking in Arabidopsis
Erratum in
-
Erratum.Plant Cell. 2022 Jan 20;34(1):700. doi: 10.1093/plcell/koab233. Plant Cell. 2022. PMID: 34689204 Free PMC article. No abstract available.
Abstract
Polycomb group (PcG) complexes ensure that every cell in an organism expresses the genes needed at a particular stage, time, or condition. However, it is still not fully understood how PcG complexes PcG-repressive complex 1 (PRC1) and PRC2 are recruited to target genes in plants. Recent findings in Arabidopsis thaliana support the notion that PRC2 recruitment is mediated by different transcription factors (TFs). However, it is unclear how all these TFs interact with PRC2 and whether they also recruit PRC1 activity. Here, by using a system to bind selected TFs to a synthetic promoter lacking the complexity of PcG target promoters in vivo, we show that while binding of the TF VIVIPAROUS1/ABSCISIC ACID-INSENSITIVE3-LIKE1 recapitulates PRC1 and PRC2 marking, the binding of other TFs only renders PRC2 marking. Interestingly, all these TFs contain an Ethylene-responsive element binding factor-associated Amphiphilic Repression (EAR) domain that triggers both HISTONE DEACETYLASE COMPLEX and PRC2 activities, connecting two different repressive mechanisms. Furthermore, we show that different TFs can have an additive effect on PRC2 activity, which may be required to maintain long-term repression of gene expression.
© The Author(s) 2021. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Figures

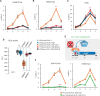
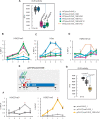
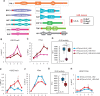


Comment in
-
An open EAR for polycomb repressive complexes.Plant Cell. 2021 Aug 31;33(8):2517-2518. doi: 10.1093/plcell/koab156. Plant Cell. 2021. PMID: 35233623 Free PMC article. No abstract available.
References
-
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M (2010) Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859 - PubMed
-
- Calonje M (2014) PRC1 marks the difference in plant PcG repression. Mol Plant 7: 459–471 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

