Computational Model-Based Estimation of Mouse Eyeball Structure From Two-Dimensional Flatmount Microscopy Images
- PMID: 34004004
- PMCID: PMC8088229
- DOI: 10.1167/tvst.10.4.25
Computational Model-Based Estimation of Mouse Eyeball Structure From Two-Dimensional Flatmount Microscopy Images
Abstract
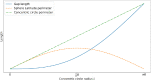
Purpose: Retinal pigment epithelial (RPE) cells serve as a supporter for the metabolism and visual function of photoreceptors and a barrier for photoreceptor protection. Morphology dynamics, spatial organization, distribution density, and growth patterns of RPE cells are important for further research on these RPE main functions. To enable such investigations within the authentic eyeball structure, a new method for estimating the three-dimensional (3D) eyeball sphere from two-dimensional tissue flatmount microscopy images was investigated.
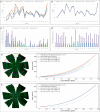
Methods: An error-correction term was formulated to compensate for the reconstruction error as a result of tissue distortions. The effect of the tissue-distortion error was evaluated by excluding partial data points from the low- and high-latitude zones. The error-correction parameter was learned automatically using a set of samples with the ground truth eyeball diameters measured with noncontact light-emitting diode micrometry at submicron accuracy and precision.
Results: The analysis showed that the error-correction term in the reconstruction model is a valid method for modeling tissue distortions in the tissue flatmount preparation steps. With the error-correction model, the average relative error of the estimated eyeball diameter was reduced from 14% to 5%, and the absolute error was reduced from 0.22 to 0.03 mm.
Conclusions: A new method for enabling RPE morphometry analysis with respect to locations on an eyeball sphere was created, an important step in increasing RPE research and eye disease diagnosis.
Translational relevance: This method enables one to derive RPE cell information from the 3D eyeball surface and helps characterize eyeball volume growth patterns under diseased conditions.
Conflict of interest statement
Disclosure:
Figures








Similar articles
-
Methodologies for analysis of patterning in the mouse RPE sheet.Mol Vis. 2015 Jan 15;21:40-60. eCollection 2015. Mol Vis. 2015. PMID: 25593512 Free PMC article.
-
MultiHeadGAN: A deep learning method for low contrast retinal pigment epithelium cell segmentation with fluorescent flatmount microscopy images.Comput Biol Med. 2022 Jul;146:105596. doi: 10.1016/j.compbiomed.2022.105596. Epub 2022 May 10. Comput Biol Med. 2022. PMID: 35617723
-
Morphometric Analysis of Retinal Pigment Epithelial Cells From C57BL/6J Mice During Aging.Invest Ophthalmol Vis Sci. 2021 Feb 1;62(2):32. doi: 10.1167/iovs.62.2.32. Invest Ophthalmol Vis Sci. 2021. PMID: 33616620 Free PMC article.
-
Relationship between Three-Dimensional Magnetic Resonance Imaging Eyeball Shape and Optic Nerve Head Morphology.Ophthalmology. 2021 Apr;128(4):532-544. doi: 10.1016/j.ophtha.2020.08.034. Epub 2020 Sep 8. Ophthalmology. 2021. PMID: 32916169
-
[Retinal pigment epithelial cell transplantation: perspective].Nippon Ganka Gakkai Zasshi. 1996 Dec;100(12):982-1006. Nippon Ganka Gakkai Zasshi. 1996. PMID: 9022310 Review. Japanese.
References
-
- Boyer MM, Poulsen GL, Nork TM. Relative contributions of the neurosensory retina and retinal pigment epithelium to macular hypofluorescence. Arch Ophthalmol. 2000; 118: 27–31. - PubMed
-
- Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005; 85: 845–881. - PubMed
-
- Liang FQ, Godley BF. Oxidative stress-induced mitochondrial DNA damage in human retinal pigment epithelial cells: a possible mechanism for RPE aging and age-related macular degeneration. Exp Eye Res. 2003; 76: 397–403. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

