Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells
- PMID: 34004174
- PMCID: PMC8116342
- DOI: 10.1016/j.celrep.2021.109179
Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells
Abstract
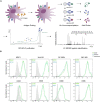
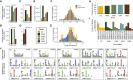
Understanding and eliciting protective immune responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is an urgent priority. To facilitate these objectives, we profile the repertoire of human leukocyte antigen class II (HLA-II)-bound peptides presented by HLA-DR diverse monocyte-derived dendritic cells pulsed with SARS-CoV-2 spike (S) protein. We identify 209 unique HLA-II-bound peptide sequences, many forming nested sets, which map to sites throughout S including glycosylated regions. Comparison of the glycosylation profile of the S protein to that of the HLA-II-bound S peptides reveals substantial trimming of glycan residues on the latter, likely induced during antigen processing. Our data also highlight the receptor-binding motif in S1 as a HLA-DR-binding peptide-rich region and identify S2-derived peptides with potential for targeting by cross-protective vaccine-elicited responses. Results from this study will aid analysis of CD4+ T cell responses in infected individuals and vaccine recipients and have application in next-generation vaccine design.
Keywords: HLA class II; HLA-II; LC-MS; SARS-CoV-2; antigen presentation; dentritic cells; glycopeptides; glycoslyation; human leukocyte antigen; immunopeptidomics.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests N.T. is directing immunopeptidomics research at Enara Bio part-time and serves on the Scientific Advisory Boards of Enara Bio and T-Cypher Bio. N.T. is consultant to Hoffman-La Roche and Grey Wolf Therapeutics. All other authors declare no conflict of interest.
Figures




Update of
-
Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells.bioRxiv [Preprint]. 2020 Aug 20:2020.08.19.255901. doi: 10.1101/2020.08.19.255901. bioRxiv. 2020. Update in: Cell Rep. 2021 May 25;35(8):109179. doi: 10.1016/j.celrep.2021.109179. PMID: 32839772 Free PMC article. Updated. Preprint.
References
-
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: Specificity, function, durability, and role in protection. Sci. Immunol. 2020;5 eabd6160. - PubMed
-
- Alves M.J., Kawahara R., Viner R., Colli W., Mattos E.C., Thaysen-Andersen M., Larsen M.R., Palmisano G. Comprehensive glycoprofiling of the epimastigote and trypomastigote stages of Trypanosoma cruzi. J. Proteomics. 2017;151:182–192. - PubMed
-
- Baum L.G., Cobb B.A. The direct and indirect effects of glycans on immune function. Glycobiology. 2017;27:619–624. - PubMed
-
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

