Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery
- PMID: 34006103
- PMCID: PMC8165695
- DOI: 10.1021/acs.jmedchem.0c02177
Re-emerging Aspartic Protease Targets: Examining Cryptococcus neoformans Major Aspartyl Peptidase 1 as a Target for Antifungal Drug Discovery
Abstract
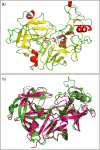
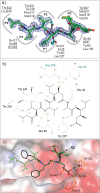
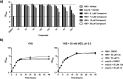
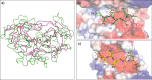
Cryptococcosis is an invasive infection that accounts for 15% of AIDS-related fatalities. Still, treating cryptococcosis remains a significant challenge due to the poor availability of effective antifungal therapies and emergence of drug resistance. Interestingly, protease inhibitor components of antiretroviral therapy regimens have shown some clinical benefits in these opportunistic infections. We investigated Major aspartyl peptidase 1 (May1), a secreted Cryptococcus neoformans protease, as a possible target for the development of drugs that act against both fungal and retroviral aspartyl proteases. Here, we describe the biochemical characterization of May1, present its high-resolution X-ray structure, and provide its substrate specificity analysis. Through combinatorial screening of 11,520 compounds, we identified a potent inhibitor of May1 and HIV protease. This dual-specificity inhibitor exhibits antifungal activity in yeast culture, low cytotoxicity, and low off-target activity against host proteases and could thus serve as a lead compound for further development of May1 and HIV protease inhibitors.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




Similar articles
-
On-Resin Assembly of Macrocyclic Inhibitors of Cryptococcus neoformans May1: A Pathway to Potent Antifungal Agents.J Med Chem. 2025 May 8;68(9):9623-9637. doi: 10.1021/acs.jmedchem.5c00396. Epub 2025 Apr 22. J Med Chem. 2025. PMID: 40262033 Free PMC article.
-
Aspartyl peptidase May1 induces host inflammatory response by altering cell wall composition in the fungal pathogen Cryptococcus neoformans.mBio. 2024 Jun 12;15(6):e0092024. doi: 10.1128/mbio.00920-24. Epub 2024 May 14. mBio. 2024. PMID: 38742885 Free PMC article.
-
Integrated Activity and Genetic Profiling of Secreted Peptidases in Cryptococcus neoformans Reveals an Aspartyl Peptidase Required for Low pH Survival and Virulence.PLoS Pathog. 2016 Dec 15;12(12):e1006051. doi: 10.1371/journal.ppat.1006051. eCollection 2016 Dec. PLoS Pathog. 2016. PMID: 27977806 Free PMC article.
-
Targeting adhesion in fungal pathogen Candida albicans.Future Med Chem. 2021 Feb;13(3):313-334. doi: 10.4155/fmc-2020-0052. Epub 2020 Jun 22. Future Med Chem. 2021. PMID: 32564615 Review.
-
Aspartic protease inhibitors as potential anti-Candida albicans drugs: impacts on fungal biology, virulence and pathogenesis.Curr Med Chem. 2011;18(16):2401-19. doi: 10.2174/092986711795843182. Curr Med Chem. 2011. PMID: 21568917 Review.
Cited by
-
On-Resin Assembly of Macrocyclic Inhibitors of Cryptococcus neoformans May1: A Pathway to Potent Antifungal Agents.J Med Chem. 2025 May 8;68(9):9623-9637. doi: 10.1021/acs.jmedchem.5c00396. Epub 2025 Apr 22. J Med Chem. 2025. PMID: 40262033 Free PMC article.
-
Genomic Diversity and Phenotypic Variation in Fungal Decomposers Involved in Bioremediation of Persistent Organic Pollutants.J Fungi (Basel). 2023 Mar 29;9(4):418. doi: 10.3390/jof9040418. J Fungi (Basel). 2023. PMID: 37108874 Free PMC article.
-
Aspartyl peptidase May1 induces host inflammatory response by altering cell wall composition in the fungal pathogen Cryptococcus neoformans.mBio. 2024 Jun 12;15(6):e0092024. doi: 10.1128/mbio.00920-24. Epub 2024 May 14. mBio. 2024. PMID: 38742885 Free PMC article.
-
The utility of Drosophila melanogaster as a fungal infection model.Front Immunol. 2024 Mar 14;15:1349027. doi: 10.3389/fimmu.2024.1349027. eCollection 2024. Front Immunol. 2024. PMID: 38550600 Free PMC article. Review.
-
Natural compounds from freshwater mussels disrupt fungal virulence determinants and influence fluconazole susceptibility in the presence of macrophages in Cryptococcus neoformans.Microbiol Spectr. 2024 Mar 5;12(3):e0284123. doi: 10.1128/spectrum.02841-23. Epub 2024 Feb 8. Microbiol Spectr. 2024. PMID: 38329361 Free PMC article.
References
-
- Kwon-Chung K. J.; Boekhout T.; Wickes B. L.; Fell J. W.. Systematics of the Genus Cryptococcus and its Type Species C. Neoformans. Cryptococcus; ASM Press, 2011; Vol. 1; pp 3–15.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

