Genomic features of rapid versus late relapse in triple negative breast cancer
- PMID: 34006255
- PMCID: PMC8130400
- DOI: 10.1186/s12885-021-08320-7
Genomic features of rapid versus late relapse in triple negative breast cancer
Abstract
Background: Triple-negative breast cancer (TNBC) is a heterogeneous disease and we have previously shown that rapid relapse of TNBC is associated with distinct sociodemographic features. We hypothesized that rapid versus late relapse in TNBC is also defined by distinct clinical and genomic features of primary tumors.
Methods: Using three publicly-available datasets, we identified 453 patients diagnosed with primary TNBC with adequate follow-up to be characterized as 'rapid relapse' (rrTNBC; distant relapse or death ≤2 years of diagnosis), 'late relapse' (lrTNBC; > 2 years) or 'no relapse' (nrTNBC: > 5 years no relapse/death). We explored basic clinical and primary tumor multi-omic data, including whole transcriptome (n = 453), and whole genome copy number and mutation data for 171 cancer-related genes (n = 317). Association of rapid relapse with clinical and genomic features were assessed using Pearson chi-squared tests, t-tests, ANOVA, and Fisher exact tests. We evaluated logistic regression models of clinical features with subtype versus two models that integrated significant genomic features.
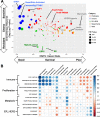
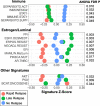
Results: Relative to nrTNBC, both rrTNBC and lrTNBC had significantly lower immune signatures and immune signatures were highly correlated to anti-tumor CD8 T-cell, M1 macrophage, and gamma-delta T-cell CIBERSORT inferred immune subsets. Intriguingly, lrTNBCs were enriched for luminal signatures. There was no difference in tumor mutation burden or percent genome altered across groups. Logistic regression mModels that incorporate genomic features significantly outperformed standard clinical/subtype models in training (n = 63 patients), testing (n = 63) and independent validation (n = 34) cohorts, although performance of all models were overall modest.
Conclusions: We identify clinical and genomic features associated with rapid relapse TNBC for further study of this aggressive TNBC subset.
Keywords: Breast Cancer; Machine learning; Triple-negative breast cancer.
Conflict of interest statement
E.P.W. has received research grants from Genentech and Roche. N.U.L. has received research grants from Genentech, Array Biopharma, GlaxoSmithKline, Kadmon and Novartis. R.W. has received research support from Acerta and Astra Zeneca and served on advisory boards for PUMA and Pfizer.
Figures



References
-
- Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. - DOI - PubMed
-
- Lin NU, Vanderplas A, Hughes ME, Theriault RL, Edge SB, Wong YN, Blayney DW, Niland JC, Winer EP, Weeks JC. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the national comprehensive Cancer network. Cancer. 2012;118(22):5463–5472. doi: 10.1002/cncr.27581. - DOI - PMC - PubMed
-
- Kassam F, Enright K, Dent R, Dranitsaris G, Myers J, Flynn C, Fralick M, Kumar R, Clemons M. Survival outcomes for patients with metastatic triple-negative breast cancer: implications for clinical practice and trial design. Clin Breast Cancer. 2009;9(1):29–33. doi: 10.3816/CBC.2009.n.005. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

