DOE JGI Metagenome Workflow
- PMID: 34006627
- PMCID: PMC8269246
- DOI: 10.1128/mSystems.00804-20
DOE JGI Metagenome Workflow
Abstract
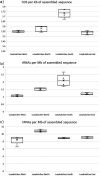
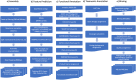
The DOE Joint Genome Institute (JGI) Metagenome Workflow performs metagenome data processing, including assembly; structural, functional, and taxonomic annotation; and binning of metagenomic data sets that are subsequently included into the Integrated Microbial Genomes and Microbiomes (IMG/M) (I.-M. A. Chen, K. Chu, K. Palaniappan, A. Ratner, et al., Nucleic Acids Res, 49:D751-D763, 2021, https://doi.org/10.1093/nar/gkaa939) comparative analysis system and provided for download via the JGI data portal (https://genome.jgi.doe.gov/portal/). This workflow scales to run on thousands of metagenome samples per year, which can vary by the complexity of microbial communities and sequencing depth. Here, we describe the different tools, databases, and parameters used at different steps of the workflow to help with the interpretation of metagenome data available in IMG and to enable researchers to apply this workflow to their own data. We use 20 publicly available sediment metagenomes to illustrate the computing requirements for the different steps and highlight the typical results of data processing. The workflow modules for read filtering and metagenome assembly are available as a workflow description language (WDL) file (https://code.jgi.doe.gov/BFoster/jgi_meta_wdl). The workflow modules for annotation and binning are provided as a service to the user community at https://img.jgi.doe.gov/submit and require filling out the project and associated metadata descriptions in the Genomes OnLine Database (GOLD) (S. Mukherjee, D. Stamatis, J. Bertsch, G. Ovchinnikova, et al., Nucleic Acids Res, 49:D723-D733, 2021, https://doi.org/10.1093/nar/gkaa983).IMPORTANCE The DOE JGI Metagenome Workflow is designed for processing metagenomic data sets starting from Illumina fastq files. It performs data preprocessing, error correction, assembly, structural and functional annotation, and binning. The results of processing are provided in several standard formats, such as fasta and gff, and can be used for subsequent integration into the Integrated Microbial Genomes and Microbiomes (IMG/M) system where they can be compared to a comprehensive set of publicly available metagenomes. As of 30 July 2020, 7,155 JGI metagenomes have been processed by the DOE JGI Metagenome Workflow. Here, we present a metagenome workflow developed at the JGI that generates rich data in standard formats and has been optimized for downstream analyses ranging from assessment of the functional and taxonomic composition of microbial communities to genome-resolved metagenomics and the identification and characterization of novel taxa. This workflow is currently being used to analyze thousands of metagenomic data sets in a consistent and standardized manner.
Keywords: IMG; JGI; SOP; annotation; assembly; binning; metagenomics.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
