A Generic, Scalable, and Rapid Time-Resolved Förster Resonance Energy Transfer-Based Assay for Antigen Detection-SARS-CoV-2 as a Proof of Concept
- PMID: 34006662
- PMCID: PMC8262888
- DOI: 10.1128/mBio.00902-21
A Generic, Scalable, and Rapid Time-Resolved Förster Resonance Energy Transfer-Based Assay for Antigen Detection-SARS-CoV-2 as a Proof of Concept
Abstract
The ongoing coronavirus disease 2019 (COVID-19) pandemic has seen an unprecedented increase in the demand for rapid and reliable diagnostic tools, leaving many laboratories scrambling for resources. We present a fast and simple assay principle for antigen detection and demonstrate its functionality by detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antigens in nasopharyngeal swabs. The method is based on the detection of SARS-CoV-2 nucleoprotein (NP) and S protein (SP) via time-resolved Förster resonance energy transfer (TR-FRET) with donor- and acceptor-labeled polyclonal anti-NP and -SP antibodies. Using recombinant proteins and cell culture-grown SARS-CoV-2, the limits of detection were established as 25 pg of NP or 20 infectious units (IU) and 875 pg of SP or 625 IU. Testing reverse transcription-PCR (RT-PCR)-positive (n = 48, with cycle threshold [CT ] values from 11 to 30) or -negative (n = 96) nasopharyngeal swabs demonstrated that the assay yielded positive results for all samples with CT values of <25 and for a single RT-PCR-negative sample. Virus isolation from the RT-PCR-positive nasopharyngeal swabs showed a strong association between the presence of infectious virus and a positive antigen test result. The NP-based assay showed 97.4% (37/38) sensitivity and 100% (10/10) specificity in comparison with virus isolation and 77.1% (37/48) sensitivity and 99.0% (95/96) specificity in comparison with SARS-CoV-2 RT-PCR. The assay is performed in a buffer that neutralizes SARS-CoV-2 infectivity, and the assay is relatively simple to set up as an "in-house" test. Here, SARS-CoV-2 served as the model pathogen, but the assay principle is applicable to other viral infections, and the test format could easily be adapted to high-throughput testing.IMPORTANCE PCR is currently the gold standard for the diagnosis of many acute infections. While PCR and its variants are highly sensitive and specific, the time from sampling to results is measured in hours at best. Antigen tests directly detect parts of the infectious agent, which may enable faster diagnosis but often at lower sensitivity and specificity. Here, we describe a technique for rapid antigen detection and demonstrate the test format's potential using SARS-CoV-2 as the model pathogen. The 10-min test, performed in a buffer that readily inactivates SARS-CoV-2, from nasopharyngeal samples identified 97.4% (37/38) of the samples from which we could isolate the virus. This suggests that the test performs well in identifying patients potentially shedding the virus. Although SARS-CoV-2 served as the model pathogen to demonstrate proof of concept, the test principle itself would be applicable to a wide variety of infectious and perhaps also noninfectious diseases.
Keywords: COVID-19; SARS-CoV-2; TR-FRET; antigen test; mix and read; rapid diagnostic test.
Copyright © 2021 Rusanen et al.
Figures
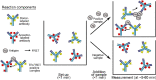
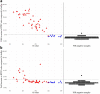
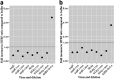
References
-
- Arons MM, Hatfield KM, Reddy SC, Kimball A, James A, Jacobs JR, Taylor J, Spicer K, Bardossy AC, Oakley LP, Tanwar S, Dyal JW, Harney J, Chisty Z, Bell JM, Methner M, Paul P, Carlson CM, McLaughlin HP, Thornburg N, Tong S, Tamin A, Tao Y, Uehara A, Harcourt J, Clark S, Brostrom-Smith C, Page LC, Kay M, Lewis J, Montgomery P, Stone ND, Clark TA, Honein MA, Duchin JS, Jernigan JA. 2020. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med 382:2081–2090. doi:10.1056/NEJMoa2008457. - DOI - PMC - PubMed
-
- Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, Boodman C, Bello A, Hedley A, Schiffman Z, Doan K, Bastien N, Li Y, Van Caeseele PG, Poliquin G. 2020. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 71:2663–2666. doi:10.1093/cid/ciaa638. - DOI - PMC - PubMed
-
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. doi:10.1038/s41586-020-2196-x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
