ESX-1-Independent Horizontal Gene Transfer by Mycobacterium tuberculosis Complex Strains
- PMID: 34006663
- PMCID: PMC8262963
- DOI: 10.1128/mBio.00965-21
ESX-1-Independent Horizontal Gene Transfer by Mycobacterium tuberculosis Complex Strains
Abstract
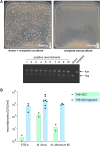
Current models of horizontal gene transfer (HGT) in mycobacteria are based on "distributive conjugal transfer" (DCT), an HGT type described in the fast-growing, saprophytic model organism Mycobacterium smegmatis, which creates genome mosaicism in resulting strains and depends on an ESX-1 type VII secretion system. In contrast, only few data on interstrain DNA transfer are available for tuberculosis-causing mycobacteria, for which chromosomal DNA transfer between two Mycobacterium canettii strains was reported, a process which, however, was not observed for Mycobacterium tuberculosis strains. Here, we have studied a wide range of human- and animal-adapted members of the Mycobacterium tuberculosis complex (MTBC) using an optimized filter-based mating assay together with three selected strains of M. canettii that acted as DNA recipients. Unlike in previous approaches, we obtained a high yield of thousands of recombinants containing transferred chromosomal DNA fragments from various MTBC donor strains, as confirmed by whole-genome sequence analysis of 38 randomly selected clones. While the genome organizations of the obtained recombinants showed mosaicisms of donor DNA fragments randomly integrated into a recipient genome backbone, reminiscent of those described as being the result of ESX-1-mediated DCT in M. smegmatis, we observed similar transfer efficiencies when ESX-1-deficient donor and/or recipient mutants were used, arguing that in tubercle bacilli, HGT is an ESX-1-independent process. These findings provide new insights into the genetic events driving the pathoevolution of M. tuberculosis and radically change our perception of HGT in mycobacteria, particularly for those species that show recombinogenic population structures despite the natural absence of ESX-1 secretion systems.IMPORTANCE Data on the bacterial sex-mediated impact on mycobacterial evolution are limited. Hence, our results presented here are of importance as they clearly demonstrate the capacity of a wide range of human- and animal-adapted Mycobacterium tuberculosis complex (MTBC) strains to transfer chromosomal DNA to selected strains of Mycobacteriumcanettii Most interestingly, we found that interstrain DNA transfer among tubercle bacilli was not dependent on a functional ESX-1 type VII secretion system, as ESX-1 deletion mutants of potential donor and/or recipient strains yielded numbers of recombinants similar to those of their respective parental strains. These results argue that HGT in tubercle bacilli is organized in a way different from that of the most widely studied Mycobacterium smegmatis model, a finding that is also relevant beyond tubercle bacilli, given that many mycobacteria, like, for example, Mycobacterium avium or Mycobacterium abscessus, are naturally devoid of an ESX-1 secretion system but show recombinogenic, mosaic-like genomic population structures.
Keywords: DNA transfer; ESX-1; Mycobacterium canettii; Mycobacterium tuberculosis; conjugation; recombinant.
Copyright © 2021 Madacki et al.
Figures



Similar articles
-
A Polymorphic Gene within the Mycobacterium smegmatis esx1 Locus Determines Mycobacterial Self-Identity and Conjugal Compatibility.mBio. 2022 Apr 26;13(2):e0021322. doi: 10.1128/mbio.00213-22. Epub 2022 Mar 17. mBio. 2022. PMID: 35297678 Free PMC article.
-
Key experimental evidence of chromosomal DNA transfer among selected tuberculosis-causing mycobacteria.Proc Natl Acad Sci U S A. 2016 Aug 30;113(35):9876-81. doi: 10.1073/pnas.1604921113. Epub 2016 Aug 15. Proc Natl Acad Sci U S A. 2016. PMID: 27528665 Free PMC article.
-
Distributive conjugal transfer in mycobacteria generates progeny with meiotic-like genome-wide mosaicism, allowing mapping of a mating identity locus.PLoS Biol. 2013 Jul;11(7):e1001602. doi: 10.1371/journal.pbio.1001602. Epub 2013 Jul 9. PLoS Biol. 2013. PMID: 23874149 Free PMC article.
-
Distributive Conjugal Transfer: New Insights into Horizontal Gene Transfer and Genetic Exchange in Mycobacteria.Microbiol Spectr. 2014 Feb;2(1):MGM2-0022-2013. doi: 10.1128/microbiolspec.MGM2-0022-2013. Microbiol Spectr. 2014. PMID: 26082110 Review.
-
Blending genomes: distributive conjugal transfer in mycobacteria, a sexier form of HGT.Mol Microbiol. 2018 Jun;108(6):601-613. doi: 10.1111/mmi.13971. Epub 2018 May 11. Mol Microbiol. 2018. PMID: 29669186 Free PMC article. Review.
Cited by
-
A Polymorphic Gene within the Mycobacterium smegmatis esx1 Locus Determines Mycobacterial Self-Identity and Conjugal Compatibility.mBio. 2022 Apr 26;13(2):e0021322. doi: 10.1128/mbio.00213-22. Epub 2022 Mar 17. mBio. 2022. PMID: 35297678 Free PMC article.
-
Genetic stability of Mycobacterium smegmatis under the stress of first-line antitubercular agents.Elife. 2024 Nov 20;13:RP96695. doi: 10.7554/eLife.96695. Elife. 2024. PMID: 39565350 Free PMC article.
-
PPE38-Secretion-Dependent Proteins of M. tuberculosis Alter NF-kB Signalling and Inflammatory Responses in Macrophages.Front Immunol. 2021 Jul 2;12:702359. doi: 10.3389/fimmu.2021.702359. eCollection 2021. Front Immunol. 2021. PMID: 34276695 Free PMC article.
-
Horizontal Gene Transfer and Drug Resistance Involving Mycobacterium tuberculosis.Antibiotics (Basel). 2023 Aug 25;12(9):1367. doi: 10.3390/antibiotics12091367. Antibiotics (Basel). 2023. PMID: 37760664 Free PMC article.
-
Crosstalk between the ancestral type VII secretion system ESX-4 and other T7SS in Mycobacterium marinum.iScience. 2021 Dec 8;25(1):103585. doi: 10.1016/j.isci.2021.103585. eCollection 2022 Jan 21. iScience. 2021. PMID: 35005535 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

