RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis
- PMID: 34006870
- PMCID: PMC8131698
- DOI: 10.1038/s41467-021-23186-w
RAS mutations drive proliferative chronic myelomonocytic leukemia via a KMT2A-PLK1 axis
Abstract
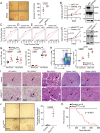
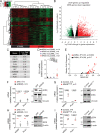
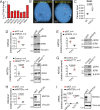
Proliferative chronic myelomonocytic leukemia (pCMML), an aggressive CMML subtype, is associated with dismal outcomes. RAS pathway mutations, mainly NRASG12D, define the pCMML phenotype as demonstrated by our exome sequencing, progenitor colony assays and a Vav-Cre-NrasG12D mouse model. Further, these mutations promote CMML transformation to acute myeloid leukemia. Using a multiomics platform and biochemical and molecular studies we show that in pCMML RAS pathway mutations are associated with a unique gene expression profile enriched in mitotic kinases such as polo-like kinase 1 (PLK1). PLK1 transcript levels are shown to be regulated by an unmutated lysine methyl-transferase (KMT2A) resulting in increased promoter monomethylation of lysine 4 of histone 3. Pharmacologic inhibition of PLK1 in RAS mutant patient-derived xenografts, demonstrates the utility of personalized biomarker-driven therapeutics in pCMML.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Coston, T. et al. Suboptimal response rates to hypomethylating agent therapy in chronic myelomonocytic leukemia; a single institutional study of 121 patients. Am. J. Hematol.94, 767–779 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

