Transformation of microbiology data into a standardised data representation using OpenEHR
- PMID: 34006956
- PMCID: PMC8131366
- DOI: 10.1038/s41598-021-89796-y
Transformation of microbiology data into a standardised data representation using OpenEHR
Abstract
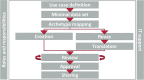
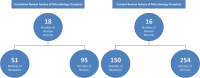
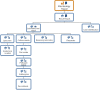
The spread of multidrug resistant organisms (MDRO) is a global healthcare challenge. Nosocomial outbreaks caused by MDRO are an important contributor to this threat. Computer-based applications facilitating outbreak detection can be essential to address this issue. To allow application reusability across institutions, the various heterogeneous microbiology data representations needs to be transformed into standardised, unambiguous data models. In this work, we present a multi-centric standardisation approach by using openEHR as modelling standard. Data models have been consented in a multicentre and international approach. Participating sites integrated microbiology reports from primary source systems into an openEHR-based data platform. For evaluation, we implemented a prototypical application, compared the transformed data with original reports and conducted automated data quality checks. We were able to develop standardised and interoperable microbiology data models. The publicly available data models can be used across institutions to transform real-life microbiology reports into standardised representations. The implementation of a proof-of-principle and quality control application demonstrated that the new formats as well as the integration processes are feasible. Holistic transformation of microbiological data into standardised openEHR based formats is feasible in a real-life multicentre setting and lays the foundation for developing cross-institutional, automated outbreak detection systems.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

