Sex-specific nicotine sensitization and imprinting of self-administration in rats inform GWAS findings on human addiction phenotypes
- PMID: 34007041
- PMCID: PMC8358005
- DOI: 10.1038/s41386-021-01027-0
Sex-specific nicotine sensitization and imprinting of self-administration in rats inform GWAS findings on human addiction phenotypes
Abstract
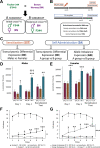
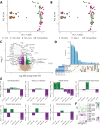
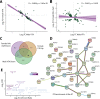
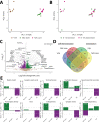
Repeated nicotine exposure leads to sensitization (SST) and enhances self-administration (SA) in rodents. However, the molecular basis of nicotine SST and SA and their biological relevance to the mounting genome-wide association study (GWAS) loci of human addictive behaviors are poorly understood. Considering a gateway drug role of nicotine, we modeled nicotine SST and SA in F1 progeny of inbred rats (F344/BN) and conducted integrative genomics analyses. We unexpectedly observed male-specific nicotine SST and a parental effect of SA only present in paternal F344 crosses. Transcriptional profiling in the ventral tegmental area (VTA) and nucleus accumbens (NAc) core and shell further revealed sex- and brain region-specific transcriptomic signatures of SST and SA. We found that genes associated with SST and SA were enriched for those related to synaptic processes, myelin sheath, and tobacco use disorder or chemdependency. Interestingly, SST-associated genes were often downregulated in male VTA but upregulated in female VTA, and strongly enriched for smoking GWAS risk variants, possibly explaining the male-specific SST. For SA, we found widespread region-specific allelic imbalance of expression (AIE), of which genes showing AIE bias toward paternal F344 alleles in NAc core were strongly enriched for SA-associated genes and for GWAS risk variants of smoking initiation, likely contributing to the parental effect of SA. Our study suggests a mechanistic link between transcriptional changes underlying the NIC SST and SA and human nicotine addiction, providing a resource for understanding the neurobiology basis of the GWAS findings on human smoking and other addictive phenotypes.
© 2021. The Author(s).
Figures

Similar articles
-
Full-gestational exposure to nicotine and ethanol augments nicotine self-administration by altering ventral tegmental dopaminergic function due to NMDA receptors in adolescent rats.J Neurochem. 2014 Mar;128(5):701-12. doi: 10.1111/jnc.12504. Epub 2013 Nov 15. J Neurochem. 2014. PMID: 24147868
-
Protection genes in nucleus accumbens shell affect vulnerability to nicotine self-administration across isogenic strains of adolescent rat.PLoS One. 2014 Jan 22;9(1):e86214. doi: 10.1371/journal.pone.0086214. eCollection 2014. PLoS One. 2014. PMID: 24465966 Free PMC article.
-
Nicotine mediates expression of genes related to antioxidant capacity and oxidative stress response in HIV-1 transgenic rat brain.J Neurovirol. 2016 Feb;22(1):114-24. doi: 10.1007/s13365-015-0375-6. Epub 2015 Aug 26. J Neurovirol. 2016. PMID: 26306689 Free PMC article.
-
Human Genetics of Addiction: New Insights and Future Directions.Curr Psychiatry Rep. 2018 Mar 5;20(2):8. doi: 10.1007/s11920-018-0873-3. Curr Psychiatry Rep. 2018. PMID: 29504045 Free PMC article. Review.
-
Tribute to: Self-administered nicotine activates the mesolimbic dopamine system through the ventral tegmental area [William Corrigall, Kathleen Coen and Laurel Adamson, Brain Res. 653 (1994) 278-284].Brain Res. 2016 Aug 15;1645:61-4. doi: 10.1016/j.brainres.2015.12.064. Epub 2016 Feb 8. Brain Res. 2016. PMID: 26867702 Review.
Cited by
-
Detection of Parent-of-Origin Effects for the Variants Associated With Behavioral Disinhibition in the MCTFR Data.Front Genet. 2022 Apr 26;13:831685. doi: 10.3389/fgene.2022.831685. eCollection 2022. Front Genet. 2022. PMID: 35559008 Free PMC article.
-
Phosphorylation of Neurofilament Light Chain in the VLO Is Correlated with Morphine-Induced Behavioral Sensitization in Rats.Int J Mol Sci. 2023 Apr 22;24(9):7709. doi: 10.3390/ijms24097709. Int J Mol Sci. 2023. PMID: 37175416 Free PMC article.
-
Transcriptomic Dysregulation in Animal Models of Attention-Deficit Hyperactivity Disorder and Nicotine Dependence Suggests Shared Neural Mechanisms.Brain Behav. 2025 Mar;15(3):e70444. doi: 10.1002/brb3.70444. Brain Behav. 2025. PMID: 40135637 Free PMC article.
-
Transcriptional characterization of cocaine withdrawal versus extinction within nucleus accumbens in male rats.Nat Commun. 2025 Mar 25;16(1):2886. doi: 10.1038/s41467-025-58151-4. Nat Commun. 2025. PMID: 40133300 Free PMC article.
References
-
- Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Soler Artigas M, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir Med. 2015;3:769–81. doi: 10.1016/S2213-2600(15)00283-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

