Coxiella burnetii and Related Tick Endosymbionts Evolved from Pathogenic Ancestors
- PMID: 34009306
- PMCID: PMC8290121
- DOI: 10.1093/gbe/evab108
Coxiella burnetii and Related Tick Endosymbionts Evolved from Pathogenic Ancestors
Abstract
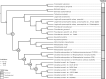
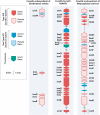
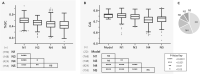
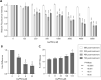
Both symbiotic and pathogenic bacteria in the family Coxiellaceae cause morbidity and mortality in humans and animals. For instance, Coxiella-like endosymbionts (CLEs) improve the reproductive success of ticks-a major disease vector, while Coxiella burnetii causes human Q fever, and uncharacterized coxiellae infect both animals and humans. To better understand the evolution of pathogenesis and symbiosis in this group of intracellular bacteria, we sequenced the genome of a CLE present in the soft tick Ornithodoros amblus (CLEOA) and compared it to the genomes of other bacteria in the order Legionellales. Our analyses confirmed that CLEOA is more closely related to C. burnetii, the human pathogen, than to CLEs in hard ticks, and showed that most clades of CLEs contain both endosymbionts and pathogens, indicating that several CLE lineages have evolved independently from pathogenic Coxiella. We also determined that the last common ancestorof CLEOA and C. burnetii was equipped to infect macrophages and that even though horizontal gene transfer (HGT) contributed significantly to the evolution of C. burnetii, most acquisition events occurred primarily in ancestors predating the CLEOA-C. burnetii divergence. These discoveries clarify the evolution of C. burnetii, which previously was assumed to have emerged when an avirulent tick endosymbiont recently gained virulence factors via HGT. Finally, we identified several metabolic pathways, including heme biosynthesis, that are likely critical to the intracellular growth of the human pathogen but not the tick symbiont, and show that the use of heme analog is a promising approach to controlling C. burnetii infections.
Keywords: Coxiella; endosymbiont; heme; pathogen; tick.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures


References
-
- Albertsen M, et al. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 31(6):533–538. - PubMed
-
- Almeida AP, et al. 2012. Coxiella symbiont in the tick Ornithodoros rostratus (Acari: argasidae). Ticks Tick Borne Dis. 3(4):203–206. - PubMed
-
- Alneberg J, et al. 2014. Binning metagenomic contigs by coverage and composition. Nat Methods. 11(11):1144–1146. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

