Converging Roles of the Aryl Hydrocarbon Receptor in Early Embryonic Development, Maintenance of Stemness, and Tissue Repair
- PMID: 34009372
- PMCID: PMC8285021
- DOI: 10.1093/toxsci/kfab050
Converging Roles of the Aryl Hydrocarbon Receptor in Early Embryonic Development, Maintenance of Stemness, and Tissue Repair
Abstract
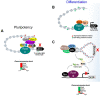
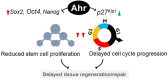
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor well-known for its adaptive role as a sensor of environmental toxicants and mediator of the metabolic detoxification of xenobiotic ligands. In addition, a growing body of experimental data has provided indisputable evidence that the AHR regulates critical functions of cell physiology and embryonic development. Recent studies have shown that the naïve AHR-that is, unliganded to xenobiotics but activated endogenously-has a crucial role in maintenance of embryonic stem cell pluripotency, tissue repair, and regulation of cancer stem cell stemness. Depending on the cellular context, AHR silences the expression of pluripotency genes Oct4 and Nanog and potentiates differentiation, whereas curtailing cellular plasticity and stemness. In these processes, AHR-mediated contextual responses and outcomes are dictated by changes of interacting partners in signaling pathways, gene networks, and cell-type-specific genomic structures. In this review, we focus on AHR-mediated changes of genomic architecture as an emerging mechanism for the AHR to regulate gene expression at the transcriptional level. Collective evidence places this receptor as a physiological hub connecting multiple biological processes whose disruption impacts on embryonic development, tissue repair, and maintenance or loss of stemness.
Keywords: Ah receptor; embryonic development; stemness; tissue repair.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society of Toxicology.All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
Similar articles
-
Pluripotency factors and Polycomb Group proteins repress aryl hydrocarbon receptor expression in murine embryonic stem cells.Stem Cell Res. 2014 Jan;12(1):296-308. doi: 10.1016/j.scr.2013.11.007. Epub 2013 Nov 16. Stem Cell Res. 2014. PMID: 24316986 Free PMC article.
-
The aryl hydrocarbon receptor promotes differentiation during mouse preimplantational embryo development.Stem Cell Reports. 2021 Sep 14;16(9):2351-2363. doi: 10.1016/j.stemcr.2021.08.002. Epub 2021 Sep 2. Stem Cell Reports. 2021. PMID: 34478649 Free PMC article.
-
Aryl hydrocarbon receptor mediates Jak2/STAT3 signaling for non-small cell lung cancer stem cell maintenance.Exp Cell Res. 2020 Nov 1;396(1):112288. doi: 10.1016/j.yexcr.2020.112288. Epub 2020 Sep 14. Exp Cell Res. 2020. PMID: 32941808
-
Regulation of Intestinal Stem Cell Stemness by the Aryl Hydrocarbon Receptor and Its Ligands.Front Immunol. 2021 Mar 10;12:638725. doi: 10.3389/fimmu.2021.638725. eCollection 2021. Front Immunol. 2021. PMID: 33777031 Free PMC article. Review.
-
The Complex Biology of the Aryl Hydrocarbon Receptor and Its Role in the Pituitary Gland.Horm Cancer. 2017 Aug;8(4):197-210. doi: 10.1007/s12672-017-0300-y. Epub 2017 Jun 20. Horm Cancer. 2017. PMID: 28634910 Free PMC article. Review.
Cited by
-
Aryl Hydrocarbon Receptor Activation Drives 2-Methoxy Estradiol Secretion in Human Trophoblast Stem Cell Development.bioRxiv [Preprint]. 2025 Jan 31:2024.08.27.609205. doi: 10.1101/2024.08.27.609205. bioRxiv. 2025. PMID: 39253430 Free PMC article. Preprint.
-
The Ah Receptor from Toxicity to Therapeutics: Report from the 5th AHR Meeting at Penn State University, USA, June 2022.Int J Mol Sci. 2023 Mar 14;24(6):5550. doi: 10.3390/ijms24065550. Int J Mol Sci. 2023. PMID: 36982624 Free PMC article.
-
From Nucleus to Organs: Insights of Aryl Hydrocarbon Receptor Molecular Mechanisms.Int J Mol Sci. 2022 Nov 29;23(23):14919. doi: 10.3390/ijms232314919. Int J Mol Sci. 2022. PMID: 36499247 Free PMC article. Review.
-
Functions of the aryl hydrocarbon receptor (AHR) beyond the canonical AHR/ARNT signaling pathway.Biochem Pharmacol. 2023 Feb;208:115371. doi: 10.1016/j.bcp.2022.115371. Epub 2022 Dec 15. Biochem Pharmacol. 2023. PMID: 36528068 Free PMC article. Review.
-
The Impact of the Aryl Hydrocarbon Receptor on Antenatal Chemical Exposure-Induced Cardiovascular-Kidney-Metabolic Programming.Int J Mol Sci. 2024 Apr 23;25(9):4599. doi: 10.3390/ijms25094599. Int J Mol Sci. 2024. PMID: 38731818 Free PMC article. Review.
References
-
- Bersten D. C., Sullivan A. E., Peet D. J., Whitelaw M. L. (2013). BHLH-PAS proteins in cancer. Nat. Rev. Cancer 13, 827–841. - PubMed
-
- Bornelöv S., Reynolds N., Xenophontos M., Gharbi S., Johnstone E., Floyd R., Ralser M., Signolet J., Loos R., Dietmann S., et al. (2018). The nucleosome remodeling and deacetylation complex modulates chromatin structure at sites of active transcription to fine-tune gene expression. Mol. Cell 71, 56–72.e4. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

