Genome-wide screening of SARS-CoV-2 infection-related genes based on the blood leukocytes sequencing data set of patients with COVID-19
- PMID: 34009691
- PMCID: PMC8242610
- DOI: 10.1002/jmv.27093
Genome-wide screening of SARS-CoV-2 infection-related genes based on the blood leukocytes sequencing data set of patients with COVID-19
Abstract
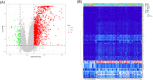
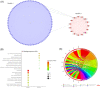
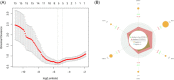
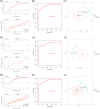
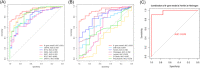
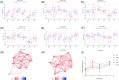
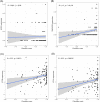
Coronavirus disease 2019 (COVID-19) is a global epidemic disease caused by a novel virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing serious adverse effects on human health. In this study, we obtained a blood leukocytes sequencing data set of COVID-19 patients from the GEO database and obtained differentially expressed genes (DEGs). We further analyzed these DEGs by protein-protein interaction analysis and Gene Ontology enrichment analysis and identified the DEGs closely related to SARS-CoV-2 infection. Then, we constructed a six-gene model (comprising IFIT3, OASL, USP18, XAF1, IFI27, and EPSTI1) by logistic regression analysis and calculated the area under the ROC curve (AUC) for the diagnosis of COVID-19. The AUC values of the training group, testing group, and entire group were 0.930, 0.914, and 0.921, respectively. The six genes were highly expressed in patients with COVID-19 and positively correlated with the expression of SARS-CoV-2 invasion-related genes (ACE2, TMPRSS2, CTSB, and CTSL). The risk score calculated by this model was also positively correlated with the expression of TMPRSS2, CTSB, and CTSL, indicating that the six genes were closely related to SARS-CoV-2 infection. In conclusion, we comprehensively analyzed the functions of DEGs in the blood leukocytes of patients with COVID-19 and constructed a six-gene model that may contribute to the development of new diagnostic and therapeutic ideas for COVID-19. Moreover, these six genes may be therapeutic targets for COVID-19.
Keywords: COVID-19; SARS-CoV-2; bioinformatics; diagnosis; leukocyte.
© 2021 Wiley Periodicals LLC.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures
Similar articles
-
Identification of common molecular signatures of SARS-CoV-2 infection and its influence on acute kidney injury and chronic kidney disease.Front Immunol. 2023 Mar 21;14:961642. doi: 10.3389/fimmu.2023.961642. eCollection 2023. Front Immunol. 2023. PMID: 37026010 Free PMC article.
-
Potential core genes associated with COVID-19 identified via weighted gene co-expression network analysis.Swiss Med Wkly. 2022 Nov 30;152:40033. doi: 10.57187/smw.2022.40033. eCollection 2022 Nov 21. Swiss Med Wkly. 2022. PMID: 36509426
-
ILRUN Downregulates ACE2 Expression and Blocks Infection of Human Cells by SARS-CoV-2.J Virol. 2021 Jul 12;95(15):e0032721. doi: 10.1128/JVI.00327-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 33963054 Free PMC article.
-
A meta-analysis of comorbidities in COVID-19: Which diseases increase the susceptibility of SARS-CoV-2 infection?Comput Biol Med. 2021 Mar;130:104219. doi: 10.1016/j.compbiomed.2021.104219. Epub 2021 Jan 16. Comput Biol Med. 2021. PMID: 33486379 Free PMC article.
-
An overview of human proteins and genes involved in SARS-CoV-2 infection.Gene. 2022 Jan 15;808:145963. doi: 10.1016/j.gene.2021.145963. Epub 2021 Sep 14. Gene. 2022. PMID: 34530086 Free PMC article. Review.
Cited by
-
Identification of COVID-19-Specific Immune Markers Using a Machine Learning Method.Front Mol Biosci. 2022 Jul 19;9:952626. doi: 10.3389/fmolb.2022.952626. eCollection 2022. Front Mol Biosci. 2022. PMID: 35928229 Free PMC article.
-
Role of E3 ubiquitin ligases and deubiquitinating enzymes in SARS-CoV-2 infection.Front Cell Infect Microbiol. 2023 Jun 9;13:1217383. doi: 10.3389/fcimb.2023.1217383. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37360529 Free PMC article. Review.
-
APOBEC3-related mutations in the spike protein-encoding region facilitate SARS-CoV-2 evolution.Heliyon. 2024 May 29;10(11):e32139. doi: 10.1016/j.heliyon.2024.e32139. eCollection 2024 Jun 15. Heliyon. 2024. PMID: 38868014 Free PMC article.
-
Functional Roles and Targets of COVID-19 in Blood Cells Determined Using Bioinformatics Analyses.Bioinform Biol Insights. 2022 Feb 23;16:11779322221080266. doi: 10.1177/11779322221080266. eCollection 2022. Bioinform Biol Insights. 2022. PMID: 35221679 Free PMC article.
-
A Survey of COVID-19 Diagnosis Using Routine Blood Tests with the Aid of Artificial Intelligence Techniques.Diagnostics (Basel). 2023 May 16;13(10):1749. doi: 10.3390/diagnostics13101749. Diagnostics (Basel). 2023. PMID: 37238232 Free PMC article. Review.
References
-
- World Health Organization. WHO Coronavirus Disease (COVID‐19) Dashboard. 2021. [Data last updated: 2021/03/22]. https://covid19.who.int.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

