Chimeric Phi29 DNA polymerase with helix-hairpin-helix motifs shows enhanced salt tolerance and replication performance
- PMID: 34009743
- PMCID: PMC8313265
- DOI: 10.1111/1751-7915.13830
Chimeric Phi29 DNA polymerase with helix-hairpin-helix motifs shows enhanced salt tolerance and replication performance
Abstract
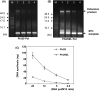
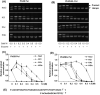
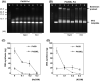
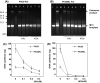
Phi29 DNA polymerase (Phi29 Pol) has been successfully applied in DNA nanoball-based sequencing, real-time DNA sequencing from single polymerase molecules and nanopore sequencing employing the sequencing by synthesis (SBS) method. Among these, polymerase-assisted nanopore sequencing technology analyses nucleotide sequences as a function of changes in electrical current. This ionic, current-based sequencing technology requires polymerases to perform replication at high salt concentrations, for example 0.3 M KCl. Nonetheless, the salt tolerance of wild-type Phi29 Pol is relatively low. Here, we fused helix-hairpin-helix (HhH)2 domains E-L (eight repeats in total) of topoisomerase V (Topo V) from the hyperthermophile Methanopyrus kandleri to the Phi29 Pol COOH terminus, designated Phi29EL DNA polymerase (Phi29EL Pol). Domain fusion increased the overall enzyme replication efficiency by fourfold. Phi29EL Pol catalysed rolling circle replication in a broader range of salt concentrations than did Phi29 Pol, extending the KCl concentration range for activity up to 0.3 M. In addition, the mutation of Glu375 to Ser or Gln increased Phi29EL Pol activity in the presence of KCl. In this work, we produced a salt-tolerant Phi29 Pol derivative by means of (HhH)2 domain insertion. The multiple advantages of this insertion make it a good substitute for Phi29 Pol, especially for use in nanopore sequencing or other circumstances that require high salt concentrations.
© 2021 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Unraveling the salt tolerance of Phi29 DNA polymerase using compartmentalized self-replication and microfluidics platform.Front Microbiol. 2023 Nov 7;14:1267196. doi: 10.3389/fmicb.2023.1267196. eCollection 2023. Front Microbiol. 2023. PMID: 38029082 Free PMC article.
-
Helix-hairpin-helix motifs confer salt resistance and processivity on chimeric DNA polymerases.Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13510-5. doi: 10.1073/pnas.202127199. Epub 2002 Oct 4. Proc Natl Acad Sci U S A. 2002. PMID: 12368475 Free PMC article.
-
Expression and functional study of VpV262 Pol, a moderately halophilic DNA polymerase from the Vibrio parahaemolyticus phage VpV262.Enzyme Microb Technol. 2020 Sep;139:109588. doi: 10.1016/j.enzmictec.2020.109588. Epub 2020 Jun 5. Enzyme Microb Technol. 2020. PMID: 32732037
-
DNA-Binding Proteins Essential for Protein-Primed Bacteriophage Φ29 DNA Replication.Front Mol Biosci. 2016 Aug 5;3:37. doi: 10.3389/fmolb.2016.00037. eCollection 2016. Front Mol Biosci. 2016. PMID: 27547754 Free PMC article. Review.
-
DNA polymerase θ (POLQ), double-strand break repair, and cancer.DNA Repair (Amst). 2016 Aug;44:22-32. doi: 10.1016/j.dnarep.2016.05.003. Epub 2016 May 14. DNA Repair (Amst). 2016. PMID: 27264557 Free PMC article. Review.
Cited by
-
Improved single-cell genome amplification by a high-efficiency phi29 DNA polymerase.Front Bioeng Biotechnol. 2023 Jun 29;11:1233856. doi: 10.3389/fbioe.2023.1233856. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 37456715 Free PMC article.
-
Engineering psychrophilic polymerase for nanopore long-read sequencing.Front Bioeng Biotechnol. 2024 Jul 1;12:1406722. doi: 10.3389/fbioe.2024.1406722. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39011153 Free PMC article.
-
Unraveling the salt tolerance of Phi29 DNA polymerase using compartmentalized self-replication and microfluidics platform.Front Microbiol. 2023 Nov 7;14:1267196. doi: 10.3389/fmicb.2023.1267196. eCollection 2023. Front Microbiol. 2023. PMID: 38029082 Free PMC article.
-
Expression and functional study of DNA polymerases from Psychrobacillus sp. BL-248-WT-3 and FJAT-21963.Front Microbiol. 2024 Nov 20;15:1501020. doi: 10.3389/fmicb.2024.1501020. eCollection 2024. Front Microbiol. 2024. PMID: 39633807 Free PMC article.
-
Strategies and procedures to generate chimeric DNA polymerases for improved applications.Appl Microbiol Biotechnol. 2024 Aug 21;108(1):445. doi: 10.1007/s00253-024-13276-2. Appl Microbiol Biotechnol. 2024. PMID: 39167106 Free PMC article. Review.
References
-
- Baba, M. , Kakue, R. , Leucht, C. , Rasor, P. , Walch, H. , Ladiges, D. , et al. (2017) Further increase in thermostability of Moloney murine leukemia virus reverse transcriptase by mutational combination. Protein Eng Des Sel 30: 551–557. - PubMed
-
- Belova, G.I. , Prasad, R. , Nazimov, I.V. , Wilson, S.H. , and Slesarev, A.I. (2002) The domain organization and properties of individual domains of DNA topoisomerase V, a type 1B topoisomerase with DNA repair activities. J Biol Chem 277: 4959–4965. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

