Liver development is restored by blastocyst complementation of HHEX knockout in mice and pigs
- PMID: 34011403
- PMCID: PMC8132445
- DOI: 10.1186/s13287-021-02348-z
Liver development is restored by blastocyst complementation of HHEX knockout in mice and pigs
Abstract
Background: There are over 17,000 patients in the US waiting to receive liver transplants, and these numbers are increasing dramatically. Significant effort is being made to obtain functional hepatocytes and liver tissue that can for therapeutic use in patients. Blastocyst complementation is a challenging, innovative technology that could fundamentally change the future of organ transplantation. It requires the knockout (KO) of genes essential for cell or organ development in early stage host embryos followed by injection of donor pluripotent stem cells (PSCs) into host blastocysts to generate chimeric offspring in which progeny of the donor cells populate the open niche to develop functional tissues and organs.
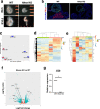
Methods: The HHEX gene is necessary for proper liver development. We engineered loss of HHEX gene expression in early mouse and pig embryos and performed intraspecies blastocyst complementation of HHEX KO embryos with eGFP-labeled PSCs in order to rescue the loss of liver development.
Results: Loss of HHEX gene expression resulted in embryonic lethality at day 10.5 in mice and produced characteristics of lethality at day 18 in pigs, with absence of liver tissue in both species. Analyses of mouse and pig HHEX KO fetuses confirmed significant loss of liver-specific gene and protein expression. Intraspecies blastocyst complementation restored liver formation and liver-specific proteins in both mouse and pig. Livers in complemented chimeric fetuses in both species were comprised of eGFP-labeled donor-derived cells and survived beyond the previously observed time of HHEX KO embryonic lethality.
Conclusions: This work demonstrates that loss of liver development in the HHEX KO can be rescued via blastocyst complementation in both mice and pigs. This complementation strategy is the first step towards generating interspecies chimeras for the goal of producing human liver cells, tissues, and potentially complete organs for clinical transplantation.
Keywords: Development; Embryo; Gene editing; Stem cells; Transplantation.
Conflict of interest statement
D.F.C. serves as C.S.O./Senior Vice President of Research and Development at Recombinetics, Inc.; W.C.L. served as C.S.O. of Regenevida a Division of Recombinetics, Inc. during the initial phase of this project; all the authors with Recombinetics, Inc. affiliation are employees and shareholders of the company; the other authors declare that they have no competing interests.
Figures



References
-
- Pettinato G, Thompson MT, Fisher RA. Human embryoid bodies to hepatocyte-like clusters: preparing for translation. Liver Res. 2017;1(2):88–95. doi: 10.1016/j.livres.2017.08.004. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

