Novel Alzheimer's disease risk variants identified based on whole-genome sequencing of APOE ε4 carriers
- PMID: 34011927
- PMCID: PMC8134477
- DOI: 10.1038/s41398-021-01412-9
Novel Alzheimer's disease risk variants identified based on whole-genome sequencing of APOE ε4 carriers
Abstract
Alzheimer's disease (AD) is a progressive neurodegenerative disease associated with a complex genetic etiology. Besides the apolipoprotein E ε4 (APOE ε4) allele, a few dozen other genetic loci associated with AD have been identified through genome-wide association studies (GWAS) conducted mainly in individuals of European ancestry. Recently, several GWAS performed in other ethnic groups have shown the importance of replicating studies that identify previously established risk loci and searching for novel risk loci. APOE-stratified GWAS have yielded novel AD risk loci that might be masked by, or be dependent on, APOE alleles. We performed whole-genome sequencing (WGS) on DNA from blood samples of 331 AD patients and 169 elderly controls of Korean ethnicity who were APOE ε4 carriers. Based on WGS data, we designed a customized AD chip (cAD chip) for further analysis on an independent set of 543 AD patients and 894 elderly controls of the same ethnicity, regardless of their APOE ε4 allele status. Combined analysis of WGS and cAD chip data revealed that SNPs rs1890078 (P = 6.64E-07) and rs12594991 (P = 2.03E-07) in SORCS1 and CHD2 genes, respectively, are novel genetic variants among APOE ε4 carriers in the Korean population. In addition, nine possible novel variants that were rare in individuals of European ancestry but common in East Asia were identified. This study demonstrates that APOE-stratified analysis is important for understanding the genetic background of AD in different populations.
Conflict of interest statement
The authors declare no competing interests.
Figures

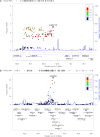
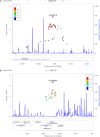
References
-
- Alzheimer’s Association Report. Alzheimer’s disease facts and figures. Alzheimers Dement.16, 391–460 10.1002/alz.12068 (2020).
-
- Kim, K. W. et al. A nationwide survey on the prevalence of dementia and mild cognitive impairment in South Korea. J. Alzheimers Dis.23, 281–291 (2011). - PubMed
-
- Cuyvers, E. & Sleegers, K. Genetic variations underlying Alzheimer’s disease: evidence from genome-wide association studies and beyond. Lancet Neurol.15, 857–868 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

