Altered endothelial dysfunction-related miRs in plasma from ME/CFS patients
- PMID: 34011981
- PMCID: PMC8134566
- DOI: 10.1038/s41598-021-89834-9
Altered endothelial dysfunction-related miRs in plasma from ME/CFS patients
Erratum in
-
Publisher Correction: Altered endothelial dysfunction-related miRs in plasma from ME/CFS patients.Sci Rep. 2021 Aug 6;11(1):16386. doi: 10.1038/s41598-021-95087-3. Sci Rep. 2021. PMID: 34362944 Free PMC article. No abstract available.
Abstract
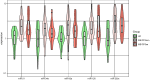
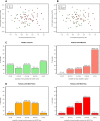
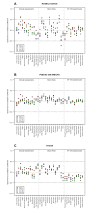
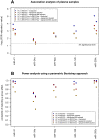
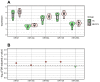
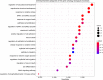
Myalgic encephalomyelitis/chronic fatigue syndrome (ME/CFS) is a complex disease characterized by unexplained debilitating fatigue. Although the etiology is unknown, evidence supports immunological abnormalities, such as persistent inflammation and immune-cell activation, in a subset of patients. Since the interplay between inflammation and vascular alterations is well-established in other diseases, endothelial dysfunction has emerged as another player in ME/CFS pathogenesis. Endothelial nitric oxide synthase (eNOS) generates nitric oxide (NO) that maintains endothelial homeostasis. eNOS is activated by silent information regulator 1 (Sirt1), an anti-inflammatory protein. Despite its relevance, no study has addressed the Sirt1/eNOS axis in ME/CFS. The interest in circulating microRNAs (miRs) as potential biomarkers in ME/CFS has increased in recent years. Accordingly, we analyze a set of miRs reported to modulate the Sirt1/eNOS axis using plasma from ME/CFS patients. Our results show that miR-21, miR-34a, miR-92a, miR-126, and miR-200c are jointly increased in ME/CFS patients compared to healthy controls. A similar finding was obtained when analyzing public miR data on peripheral blood mononuclear cells. Bioinformatics analysis shows that endothelial function-related signaling pathways are associated with these miRs, including oxidative stress and oxygen regulation. Interestingly, histone deacetylase 1, a protein responsible for epigenetic regulations, represented the most relevant node within the network. In conclusion, our study provides a basis to find endothelial dysfunction-related biomarkers and explore novel targets in ME/CFS.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

