Arylmethylene hydrazine derivatives containing 1,3-dimethylbarbituric moiety as novel urease inhibitors
- PMID: 34012008
- PMCID: PMC8134453
- DOI: 10.1038/s41598-021-90104-x
Arylmethylene hydrazine derivatives containing 1,3-dimethylbarbituric moiety as novel urease inhibitors
Abstract
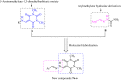
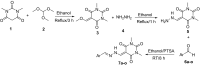
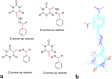
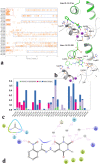
A new series of arylmethylene hydrazine derivatives bearing 1,3-dimethylbarbituric moiety 7a-o were designed, synthesized, and evaluated for their in vitro urease inhibitory activity. All the title compounds displayed high anti-urease activity, with IC50 values in the range of 0.61 ± 0.06-4.56 ± 0.18 µM as compared to the two standard inhibitors hydroxyurea (IC50 = 100 ± 0.15 μM) and thiourea (IC50 = 23 ± 1.7 μM). Among the synthesized compounds, compound 7h with 2-nitro benzylidene group was found to be the most potent compound. Kinetic study of this compound revealed that it is a mix-mode inhibitor against urease. Evaluation of the interaction modes of the synthesized compounds in urease active site by molecular modeling revealed that that compounds with higher urease inhibitor activity (7h, 7m, 7c, 7l, 7i, and 7o, with IC50 of 0.61, 0.86, 1.2, 1.34, 1.33, 1.94 μM, respectively) could interact with higher number of residues, specially Arg609, Cys592 (as part of urease active site flap) and showed higher computed free energy, while compounds with lower urease activity (7f, 7n, 7g, and 7a with IC50 of 3.56, 4.56, 3.62 and 4.43 μM, respectively) and could not provide the proper interaction with Arg609, and Cys592 as the key interacting residues along with lower free binding energy. MD investigation revealed compound 7h interacted with Arg609 and Cys592 which are of the key residues at the root part of mobile flap covering the active site. Interacting with the mentioned residue for a significant amount of time, affects the flexibility of the mobile flap covering the active site and causes inhibition of the ureolytic activity. Furthermore, in silico physico-chemical study of compounds 7a-o predicted that all these compounds are drug-likeness with considerable orally availability.
Conflict of interest statement
The authors declare no competing interests.
Figures

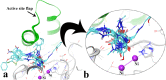
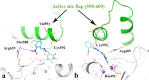
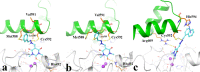
Similar articles
-
Novel N,N-dimethylbarbituric-pyridinium derivatives as potent urease inhibitors: Synthesis, in vitro, and in silico studies.Bioorg Chem. 2020 Jan;95:103529. doi: 10.1016/j.bioorg.2019.103529. Epub 2019 Dec 20. Bioorg Chem. 2020. PMID: 31884139
-
Molecular hybrids of substituted phenylcarbamoylpiperidine and 1,2,4-triazole methylacetamide as potent 15-LOX inhibitors: Design, synthesis, DFT calculations and molecular docking studies.Bioorg Chem. 2024 Feb;143:106984. doi: 10.1016/j.bioorg.2023.106984. Epub 2023 Nov 23. Bioorg Chem. 2024. PMID: 38056389
-
Design and synthesis of new N-thioacylated ciprofloxacin derivatives as urease inhibitors with potential antibacterial activity.Sci Rep. 2022 Aug 15;12(1):13827. doi: 10.1038/s41598-022-17993-4. Sci Rep. 2022. PMID: 35970866 Free PMC article.
-
Synthesis and Evaluation of 1,3,5-Triaryl-2-Pyrazoline Derivatives as Potent Dual Inhibitors of Urease and α-Glucosidase Together with Their Cytotoxic, Molecular Modeling and Drug-Likeness Studies.ACS Omega. 2022 Jan 20;7(4):3775-3795. doi: 10.1021/acsomega.1c06694. eCollection 2022 Feb 1. ACS Omega. 2022. PMID: 35128286 Free PMC article.
-
Investigations of p-tolyloxy-1,3,4-oxadiazole propionamides as soybean 15-lipoxygenase inhibitors in comforting with in vitro and in silico studies.J Biomol Struct Dyn. 2023;41(24):15549-15568. doi: 10.1080/07391102.2023.2190807. Epub 2023 Mar 22. J Biomol Struct Dyn. 2023. PMID: 36946200
Cited by
-
[1,2,4]triazolo[3,4-b][1,3,4]thiadiazole derivatives as new therapeutic candidates against urease positive microorganisms: design, synthesis, pharmacological evaluations, and in silico studies.Sci Rep. 2023 Jun 22;13(1):10136. doi: 10.1038/s41598-023-37203-z. Sci Rep. 2023. PMID: 37349372 Free PMC article.
-
Anti-urease therapy: a targeted approach to mitigating antibiotic resistance in Helicobacter pylori while preserving the gut microflora.Gut Pathog. 2025 May 28;17(1):37. doi: 10.1186/s13099-025-00708-1. Gut Pathog. 2025. PMID: 40437630 Free PMC article. Review.
-
Synthesis, Urease Inhibition, Molecular Docking, and Optical Analysis of a Symmetrical Schiff Base and Its Selected Metal Complexes.Molecules. 2024 Oct 16;29(20):4899. doi: 10.3390/molecules29204899. Molecules. 2024. PMID: 39459267 Free PMC article.
-
Synthetic Transformation of 2-{2-Fluoro[1,1'-biphenyl]-4-yl} Propanoic Acid into Hydrazide-Hydrazone Derivatives: In Vitro Urease Inhibition and In Silico Study.ACS Omega. 2022 Nov 30;7(49):45077-45087. doi: 10.1021/acsomega.2c05498. eCollection 2022 Dec 13. ACS Omega. 2022. PMID: 36530251 Free PMC article.
-
Efficient synthesis of novel phenanthroline-dimedone derivatives using Pd@HQBI-SPION as a versatile palladium-immobilized catalyst.Sci Rep. 2024 Nov 1;14(1):26325. doi: 10.1038/s41598-024-76221-3. Sci Rep. 2024. PMID: 39487194 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

