In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels
- PMID: 34012094
- PMCID: PMC8352781
- DOI: 10.1038/s41587-021-00933-4
In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels
Abstract
Most known pathogenic point mutations in humans are C•G to T•A substitutions, which can be directly repaired by adenine base editors (ABEs). In this study, we investigated the efficacy and safety of ABEs in the livers of mice and cynomolgus macaques for the reduction of blood low-density lipoprotein (LDL) levels. Lipid nanoparticle-based delivery of mRNA encoding an ABE and a single-guide RNA targeting PCSK9, a negative regulator of LDL, induced up to 67% editing (on average, 61%) in mice and up to 34% editing (on average, 26%) in macaques. Plasma PCSK9 and LDL levels were stably reduced by 95% and 58% in mice and by 32% and 14% in macaques, respectively. ABE mRNA was cleared rapidly, and no off-target mutations in genomic DNA were found. Re-dosing in macaques did not increase editing, possibly owing to the detected humoral immune response to ABE upon treatment. These findings support further investigation of ABEs to treat patients with monogenic liver diseases.
© 2021. The Author(s).
Conflict of interest statement
P.J.C.L., M.D., Y.K.T. and S.C.S. are employees of Acuitas Therapeutics. K.H., J.A.W., D.V.V. and A.P.K. are employees and shareholders of Synthego Corporation. D. Weissman is named on patents that describe the use of nucleoside-modified mRNA as a platform to deliver therapeutic proteins. M.J., G.S. and L.V. are named on patents and patent applications related to CRISPR–Cas technologies. The remaining authors declare no competing financial interests.
Figures
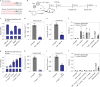
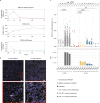

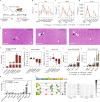
Comment in
-
Pushing the envelope with PCSK9.Nat Rev Drug Discov. 2021 Jul;20(7):506. doi: 10.1038/d41573-021-00093-9. Nat Rev Drug Discov. 2021. PMID: 34035481 No abstract available.
-
Base editing takes a shot at disease in non-human primates.Nat Rev Genet. 2021 Aug;22(8):479. doi: 10.1038/s41576-021-00382-4. Nat Rev Genet. 2021. PMID: 34045716 No abstract available.
-
Therapeutic base editing in the adult liver.Nat Rev Gastroenterol Hepatol. 2021 Sep;18(9):597-598. doi: 10.1038/s41575-021-00491-9. Nat Rev Gastroenterol Hepatol. 2021. PMID: 34285418 No abstract available.
References
-
- van den Bosch, M., Lohman, P. H. M. & Pastink, A. DNA double-strand break repair by homologous recombination. Biol. Chem.10.1515/BC.2002.095 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

