Interpreting type 1 diabetes risk with genetics and single-cell epigenomics
- PMID: 34012112
- PMCID: PMC10560508
- DOI: 10.1038/s41586-021-03552-w
Interpreting type 1 diabetes risk with genetics and single-cell epigenomics
Abstract
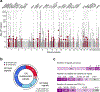
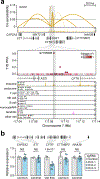
Genetic risk variants that have been identified in genome-wide association studies of complex diseases are primarily non-coding1. Translating these risk variants into mechanistic insights requires detailed maps of gene regulation in disease-relevant cell types2. Here we combined two approaches: a genome-wide association study of type 1 diabetes (T1D) using 520,580 samples, and the identification of candidate cis-regulatory elements (cCREs) in pancreas and peripheral blood mononuclear cells using single-nucleus assay for transposase-accessible chromatin with sequencing (snATAC-seq) of 131,554 nuclei. Risk variants for T1D were enriched in cCREs that were active in T cells and other cell types, including acinar and ductal cells of the exocrine pancreas. Risk variants at multiple T1D signals overlapped with exocrine-specific cCREs that were linked to genes with exocrine-specific expression. At the CFTR locus, the T1D risk variant rs7795896 mapped to a ductal-specific cCRE that regulated CFTR; the risk allele reduced transcription factor binding, enhancer activity and CFTR expression in ductal cells. These findings support a role for the exocrine pancreas in the pathogenesis of T1D and highlight the power of large-scale genome-wide association studies and single-cell epigenomics for understanding the cellular origins of complex disease.
Figures












References
-
- Katsarou A. et al. Type 1 diabetes mellitus. Nat. Rev. Dis. Primer 3, 17016 (2017). - PubMed
ADDITIONAL REFERENCES
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

