Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection
- PMID: 34012115
- PMCID: PMC7610904
- DOI: 10.1038/s41586-021-03566-4
Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection
Abstract
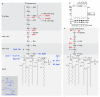
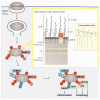
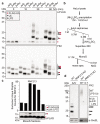
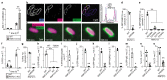
Ubiquitylation is a widespread post-translational protein modification in eukaryotes and marks bacteria that invade the cytosol as cargo for antibacterial autophagy1-3. The identity of the ubiquitylated substrate on bacteria is unknown. Here we show that the ubiquitin coat on Salmonella that invade the cytosol is formed through the ubiquitylation of a non-proteinaceous substrate, the lipid A moiety of bacterial lipopolysaccharide (LPS), by the E3 ubiquitin ligase ring finger protein 213 (RNF213). RNF213 is a risk factor for moyamoya disease4,5, which is a progressive stenosis of the supraclinoid internal carotid artery that causes stroke (especially in children)6,7. RNF213 restricts the proliferation of cytosolic Salmonella and is essential for the generation of the bacterial ubiquitin coat, both directly (through the ubiquitylation of LPS) and indirectly (through the recruitment of LUBAC, which is a downstream E3 ligase that adds M1-linked ubiquitin chains onto pre-existing ubiquitin coats8). In cells that lack RNF213, bacteria do not attract ubiquitin-dependent autophagy receptors or induce antibacterial autophagy. The ubiquitylation of LPS on Salmonella that invade the cytosol requires the dynein-like core of RNF213, but not its RING domain. Instead, ubiquitylation of LPS relies on an RZ finger in the E3 shell. We conclude that ubiquitylation extends beyond protein substrates and that ubiquitylation of LPS triggers cell-autonomous immunity, and we postulate that non-proteinaceous substances other than LPS may also become ubiquitylated.
Conflict of interest statement
The authors declare no competing interests.
Figures





Comment in
-
Host ubiquitin protein tags lipid to fight bacteria.Nature. 2021 Jun;594(7861):28-29. doi: 10.1038/d41586-021-01267-6. Nature. 2021. PMID: 34012123 No abstract available.
-
Bacterial lipids earmarked with ubiquitin for pathogen clearance.Mol Cell. 2021 Jul 1;81(13):2690-2692. doi: 10.1016/j.molcel.2021.06.012. Mol Cell. 2021. PMID: 34214444
References
-
- Perrin AJ, Jiang X, Birmingham CL, So NSY, Brumell JH. Recognition of 666 Bacteria in the Cytosol of Mammalian Cells by the Ubiquitin System. Curr Biol. 2004;14:806–811. - PubMed
-
- Kamada F, et al. A genome-wide association study identifies RNF213 as the first Moyamoya disease gene. J Hum Genet. 2011;56:34–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

