This is a preprint.
In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains
- PMID: 34013259
- PMCID: PMC8132254
- DOI: 10.21203/rs.3.rs-448370/v1
In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains
Update in
-
In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains.Nature. 2021 Aug;596(7870):103-108. doi: 10.1038/s41586-021-03720-y. Epub 2021 Jun 21. Nature. 2021. PMID: 34153975 Free PMC article.
Abstract
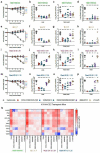
Rapidly-emerging variants jeopardize antibody-based countermeasures against SARS-CoV-2. While recent cell culture experiments have demonstrated loss of potency of several anti-spike neutralizing antibodies against SARS-CoV-2 variant strains1-3, the in vivo significance of these results remains uncertain. Here, using a panel of monoclonal antibodies (mAbs) corresponding to many in advanced clinical development by Vir Biotechnology, AbbVie, AstraZeneca, Regeneron, and Lilly we report the impact on protection in animals against authentic SARS-CoV-2 variants including WA1/2020 strains, a B.1.1.7 isolate, and chimeric strains with South African (B.1.351) or Brazilian (B.1.1.28) spike genes. Although some individual mAbs showed reduced or abrogated neutralizing activity against B.1.351 and B.1.1.28 viruses with E484K spike protein mutations in cell culture, low prophylactic doses of mAb combinations protected against infection in K18-hACE2 transgenic mice, 129S2 immunocompetent mice, and hamsters without emergence of resistance. Two exceptions were mAb LY-CoV555 monotherapy which lost all protective activity in vivo, and AbbVie 2B04/47D11, which showed partial loss of activity. When administered after infection as therapy, higher doses of mAb cocktails protected in vivo against viruses displaying a B.1.351 spike gene. Thus, many, but not all, of the antibody products with Emergency Use Authorization should retain substantial efficacy against the prevailing SARS-CoV-2 variant strains.
Figures




Similar articles
-
In vivo monoclonal antibody efficacy against SARS-CoV-2 variant strains.Nature. 2021 Aug;596(7870):103-108. doi: 10.1038/s41586-021-03720-y. Epub 2021 Jun 21. Nature. 2021. PMID: 34153975 Free PMC article.
-
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies.Res Sq [Preprint]. 2021 Dec 27:rs.3.rs-1175516. doi: 10.21203/rs.3.rs-1175516/v1. Res Sq. 2021. Update in: Nat Med. 2022 Mar;28(3):490-495. doi: 10.1038/s41591-021-01678-y. PMID: 34981042 Free PMC article. Updated. Preprint.
-
An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies.Nat Med. 2022 Mar;28(3):490-495. doi: 10.1038/s41591-021-01678-y. Epub 2022 Jan 19. Nat Med. 2022. PMID: 35046573 Free PMC article.
-
SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies.Res Sq [Preprint]. 2021 Feb 10:rs.3.rs-228079. doi: 10.21203/rs.3.rs-228079/v1. Res Sq. 2021. Update in: Nat Med. 2021 Apr;27(4):717-726. doi: 10.1038/s41591-021-01294-w. PMID: 33594356 Free PMC article. Updated. Preprint.
-
Susceptibility of SARS-CoV-2 Omicron Variants to Therapeutic Monoclonal Antibodies: Systematic Review and Meta-analysis.Microbiol Spectr. 2022 Aug 31;10(4):e0092622. doi: 10.1128/spectrum.00926-22. Epub 2022 Jun 14. Microbiol Spectr. 2022. PMID: 35700134 Free PMC article.
References
-
- Wang P., et al. Antibody Resistance of SARS-CoV-2 Variants B. 1.351 and B.1.1.7. Nature(2021). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

