This is a preprint.
SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity
- PMID: 34013273
- PMCID: PMC8132231
- DOI: 10.1101/2021.05.09.443331
SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity
Update in
-
SARS-CoV-2 ferritin nanoparticle vaccines elicit broad SARS coronavirus immunogenicity.Cell Rep. 2021 Dec 21;37(12):110143. doi: 10.1016/j.celrep.2021.110143. Epub 2021 Dec 8. Cell Rep. 2021. PMID: 34919799 Free PMC article.
Abstract
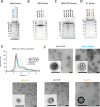
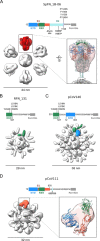
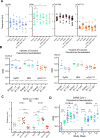
The need for SARS-CoV-2 next-generation vaccines has been highlighted by the rise of variants of concern (VoC) and the long-term threat of other coronaviruses. Here, we designed and characterized four categories of engineered nanoparticle immunogens that recapitulate the structural and antigenic properties of prefusion Spike (S), S1 and RBD. These immunogens induced robust S-binding, ACE2-inhibition, and authentic and pseudovirus neutralizing antibodies against SARS-CoV-2 in mice. A Spike-ferritin nanoparticle (SpFN) vaccine elicited neutralizing titers more than 20-fold higher than convalescent donor serum, following a single immunization, while RBD-Ferritin nanoparticle (RFN) immunogens elicited similar responses after two immunizations. Passive transfer of IgG purified from SpFN- or RFN-immunized mice protected K18-hACE2 transgenic mice from a lethal SARS-CoV-2 virus challenge. Furthermore, SpFN- and RFN-immunization elicited ACE2 blocking activity and neutralizing ID50 antibody titers >2,000 against SARS-CoV-1, along with high magnitude neutralizing titers against major VoC. These results provide design strategies for pan-coronavirus vaccine development.
Highlights: Iterative structure-based design of four Spike-domain Ferritin nanoparticle classes of immunogensSpFN-ALFQ and RFN-ALFQ immunization elicits potent neutralizing activity against SARS-CoV-2, variants of concern, and SARS-CoV-1Passively transferred IgG from immunized C57BL/6 mice protects K18-hACE2 mice from lethal SARS-CoV-2 challenge.
Conflict of interest statement
DECLARATIONS OF INTERESTS
M.G.J. and K.M. are named as inventors on International Patent Application No. WO/2021/21405 entitled “Vaccines against SARS-CoV-2 and other coronaviruses.” M.G.J. is named as an inventor on International Patent Application No. WO/2018/081318 and U.S. patent 10,960,070 entitled “Prefusion Coronavirus Spike Proteins and Their Use.” Z.B. is named as an inventor on U.S. patent 10,434,167 entitled “Non-toxic adjuvant formulation comprising a monophosphoryl lipid A (MPLA)-containing liposome composition and a saponin.” Z.B. and G.R.M are named inventors on “Compositions And Methods For Vaccine Delivery”, US Patent Application: 16/607,917. M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals and Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology and Emergent BioSolutions. S.R., P.M.M., and M.T.E. are employees of AstraZeneca and currently hold AstraZeneca stock or stock options. Zoltan Beck is currently employed at Pfizer.
Figures




References
-
- Barnes C.O., West A.P. Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. (2020). Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell 182, 828–842 e816. - PMC - PubMed
-
- Boyoglu-Barnum S., Ellis D., Gillespie R.A., Hutchinson G.B., Park Y.-J., Moin S.M., Acton O., Ravichandran R., Murphy M., Pettie D., et al. (2020). Elicitation of broadly protective immunity to influenza by multivalent hemagglutinin nanoparticle vaccines. bioRxiv, 2020.2005.2030.125179.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
