Molecular Epidemiology and Evolutionary Trajectory of Emerging Echovirus 30, Europe
- PMID: 34013874
- PMCID: PMC8153861
- DOI: 10.3201/eid2706.203096
Molecular Epidemiology and Evolutionary Trajectory of Emerging Echovirus 30, Europe
Abstract
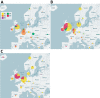
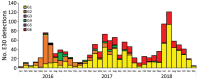
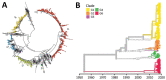
In 2018, an upsurge in echovirus 30 (E30) infections was reported in Europe. We conducted a large-scale epidemiologic and evolutionary study of 1,329 E30 strains collected in 22 countries in Europe during 2016-2018. Most E30 cases affected persons 0-4 years of age (29%) and 25-34 years of age (27%). Sequences were divided into 6 genetic clades (G1-G6). Most (53%) sequences belonged to G1, followed by G6 (23%), G2 (17%), G4 (4%), G3 (0.3%), and G5 (0.2%). Each clade encompassed unique individual recombinant forms; G1 and G4 displayed >2 unique recombinant forms. Rapid turnover of new clades and recombinant forms occurred over time. Clades G1 and G6 dominated in 2018, suggesting the E30 upsurge was caused by emergence of 2 distinct clades circulating in Europe. Investigation into the mechanisms behind the rapid turnover of E30 is crucial for clarifying the epidemiology and evolution of these enterovirus infections.
Keywords: Europe; European Non-Polio Enterovirus Network; Molecular epidemiology; Nextstrain; echovirus 30; enterovirus; epidemiological monitoring; evolutionary trajectory; genetic recombination; meningitis/encephalitis; neurological manifestations; viruses; whole-genome sequencing.
Figures


References
-
- Oberste MS, Maher K, Kennett ML, Campbell JJ, Carpenter MS, Schnurr D, et al. Molecular epidemiology and genetic diversity of echovirus type 30 (E30): genotypes correlate with temporal dynamics of E30 isolation. J Clin Microbiol. 1999;37:3928–33. 10.1128/JCM.37.12.3928-3933.1999 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

