A Bivalve Biomineralization Toolbox
- PMID: 34014311
- PMCID: PMC8382897
- DOI: 10.1093/molbev/msab153
A Bivalve Biomineralization Toolbox
Abstract
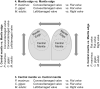
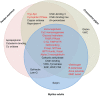
Mollusc shells are a result of the deposition of crystalline and amorphous calcite catalyzed by enzymes and shell matrix proteins (SMP). Developing a detailed understanding of bivalve mollusc biomineralization pathways is complicated not only by the multiplicity of shell forms and microstructures in this class, but also by the evolution of associated proteins by domain co-option and domain shuffling. In spite of this, a minimal biomineralization toolbox comprising proteins and protein domains critical for shell production across species has been identified. Using a matched pair design to reduce experimental noise from inter-individual variation, combined with damage-repair experiments and a database of biomineralization SMPs derived from published works, proteins were identified that are likely to be involved in shell calcification. Eighteen new, shared proteins likely to be involved in the processes related to the calcification of shells were identified by the analysis of genes expressed during repair in Crassostrea gigas, Mytilus edulis, and Pecten maximus. Genes involved in ion transport were also identified as potentially involved in calcification either via the maintenance of cell acid-base balance or transport of critical ions to the extrapallial space, the site of shell assembly. These data expand the number of candidate biomineralization proteins in bivalve molluscs for future functional studies and define a minimal functional protein domain set required to produce solid microstructures from soluble calcium carbonate. This is important for understanding molluscan shell evolution, the likely impacts of environmental change on biomineralization processes, materials science, and biomimicry research.
Keywords: Crassostrea gigas; Mytilus edulis; Pecten maximus; biomineralization; damage-repair; transcriptomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



Similar articles
-
Characterization of the mantle transcriptome in bivalves: Pecten maximus, Mytilus edulis and Crassostrea gigas.Mar Genomics. 2016 Jun;27:9-15. doi: 10.1016/j.margen.2016.04.003. Epub 2016 May 7. Mar Genomics. 2016. PMID: 27160853
-
Transcriptomic analysis of shell repair and biomineralization in the blue mussel, Mytilus edulis.BMC Genomics. 2021 Jun 10;22(1):437. doi: 10.1186/s12864-021-07751-7. BMC Genomics. 2021. PMID: 34112105 Free PMC article.
-
Insights from the Shell Proteome: Biomineralization to Adaptation.Mol Biol Evol. 2017 Jan;34(1):66-77. doi: 10.1093/molbev/msw219. Epub 2016 Oct 15. Mol Biol Evol. 2017. PMID: 27744410 Free PMC article.
-
Deciphering mollusc shell production: the roles of genetic mechanisms through to ecology, aquaculture and biomimetics.Biol Rev Camb Philos Soc. 2020 Dec;95(6):1812-1837. doi: 10.1111/brv.12640. Epub 2020 Jul 31. Biol Rev Camb Philos Soc. 2020. PMID: 32737956 Review.
-
Deciphering the molecular toolkit: regulatory elements governing shell biomineralization in marine molluscs.Integr Zool. 2025 May;20(3):448-464. doi: 10.1111/1749-4877.12876. Epub 2024 Jul 18. Integr Zool. 2025. PMID: 39030865 Review.
Cited by
-
Multi-omic insights into the formation and evolution of a novel shell microstructure in oysters.BMC Biol. 2023 Sep 29;21(1):204. doi: 10.1186/s12915-023-01706-y. BMC Biol. 2023. PMID: 37775818 Free PMC article.
-
Tracing the evolution of tissue inhibitor of metalloproteinases in Metazoa with the Pteria penguin genome.iScience. 2023 Nov 25;27(1):108579. doi: 10.1016/j.isci.2023.108579. eCollection 2024 Jan 19. iScience. 2023. PMID: 38161420 Free PMC article.
-
Physiological Roles of Serotonin in Bivalves: Possible Interference by Environmental Chemicals Resulting in Neuroendocrine Disruption.Front Endocrinol (Lausanne). 2022 Feb 25;13:792589. doi: 10.3389/fendo.2022.792589. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35282445 Free PMC article. Review.
-
Transcriptomic, Proteomic, and Functional Assays Underline the Dual Role of Extrapallial Hemocytes in Immunity and Biomineralization in the Hard Clam Mercenaria mercenaria.Front Immunol. 2022 Feb 22;13:838530. doi: 10.3389/fimmu.2022.838530. eCollection 2022. Front Immunol. 2022. PMID: 35273613 Free PMC article.
-
Metabolic profiling of Mytilus coruscus mantle in response of shell repairing under acute acidification.PLoS One. 2023 Oct 27;18(10):e0293565. doi: 10.1371/journal.pone.0293565. eCollection 2023. PLoS One. 2023. PMID: 37889901 Free PMC article.
References
-
- Aguilera F, McDougall C, Degnan BM.. 2014. Evolution of the tyrosinase gene family in bivalve molluscs: independent expansion of the mantle gene repertoire. Acta Biomater. 10(9):3855–3865. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

