HAND2 is a novel obesity-linked adipogenic transcription factor regulated by glucocorticoid signalling
- PMID: 34014371
- PMCID: PMC8245394
- DOI: 10.1007/s00125-021-05470-y
HAND2 is a novel obesity-linked adipogenic transcription factor regulated by glucocorticoid signalling
Abstract
Aims/hypothesis: Adipocytes are critical cornerstones of energy metabolism. While obesity-induced adipocyte dysfunction is associated with insulin resistance and systemic metabolic disturbances, adipogenesis, the formation of new adipocytes and healthy adipose tissue expansion are associated with metabolic benefits. Understanding the molecular mechanisms governing adipogenesis is of great clinical potential to efficiently restore metabolic health in obesity. Here we investigate the role of heart and neural crest derivatives-expressed 2 (HAND2) in adipogenesis.
Methods: Human white adipose tissue (WAT) was collected from two cross-sectional studies of 318 and 96 individuals. In vitro, for mechanistic experiments we used primary adipocytes from humans and mice as well as human multipotent adipose-derived stem (hMADS) cells. Gene silencing was performed using siRNA or genetic inactivation in primary adipocytes from loxP and or tamoxifen-inducible Cre-ERT2 mouse models with Cre-encoding mRNA or tamoxifen, respectively. Adipogenesis and adipocyte metabolism were measured by Oil Red O staining, quantitative PCR (qPCR), microarray, glucose uptake assay, western blot and lipolysis assay. A combinatorial RNA sequencing (RNAseq) and ChIP qPCR approach was used to identify target genes regulated by HAND2. In vivo, we created a conditional adipocyte Hand2 deletion mouse model using Cre under control of the Adipoq promoter (Hand2AdipoqCre) and performed a large panel of metabolic tests.
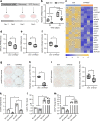
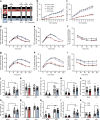
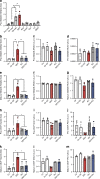
Results: We found that HAND2 is an obesity-linked white adipocyte transcription factor regulated by glucocorticoids that was necessary but insufficient for adipocyte differentiation in vitro. In a large cohort of humans, WAT HAND2 expression was correlated to BMI. The HAND2 gene was enriched in white adipocytes compared with brown, induced early in differentiation and responded to dexamethasone (DEX), a typical glucocorticoid receptor (GR, encoded by NR3C1) agonist. Silencing of NR3C1 in hMADS cells or deletion of GR in a transgenic conditional mouse model results in diminished HAND2 expression, establishing that adipocyte HAND2 is regulated by glucocorticoids via GR in vitro and in vivo. Furthermore, we identified gene clusters indirectly regulated by the GR-HAND2 pathway. Interestingly, silencing of HAND2 impaired adipocyte differentiation in hMADS and primary mouse adipocytes. However, a conditional adipocyte Hand2 deletion mouse model using Cre under control of the Adipoq promoter did not mirror these effects on adipose tissue differentiation, indicating that HAND2 was required at stages prior to Adipoq expression.
Conclusions/interpretation: In summary, our study identifies HAND2 as a novel obesity-linked adipocyte transcription factor, highlighting new mechanisms of GR-dependent adipogenesis in humans and mice.
Data availability: Array data have been submitted to the GEO database at NCBI (GSE148699).
Keywords: Adipocytes; Dexamethasone; Differentiation; Glucocorticoid receptor; HAND2; Human adipose tissue; Mesenchymal stem cells; Obesity; Transcription factor; hMADS.
Figures




References
-
- Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J Biol Chem. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. - DOI - PMC - PubMed
-
- Buren J, Liu H, Jensen J, Eriksson J (2002) Dexamethasone impairs insulin signalling and glucose transport by depletion of insulin receptor substrate-1, phosphatidylinositol 3-kinase and protein kinase B in primary cultured rat adipocytes. Eur J Endocrinol 419–429. 10.1530/eje.0.1460419 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

