Homeobox Gene Six3 is Required for the Differentiation of D2-Type Medium Spiny Neurons
- PMID: 34014554
- PMCID: PMC8275777
- DOI: 10.1007/s12264-021-00698-5
Homeobox Gene Six3 is Required for the Differentiation of D2-Type Medium Spiny Neurons
Abstract
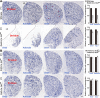
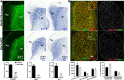
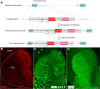
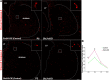
Medium spiny neurons (MSNs) in the striatum, which can be divided into D1 and D2 MSNs, originate from the lateral ganglionic eminence (LGE). Previously, we reported that Six3 is a downstream target of Sp8/Sp9 in the transcriptional regulatory cascade of D2 MSN development and that conditionally knocking out Six3 leads to a severe loss of D2 MSNs. Here, we showed that Six3 mainly functions in D2 MSN precursor cells and gradually loses its function as D2 MSNs mature. Conditional deletion of Six3 had little effect on cell proliferation but blocked the differentiation of D2 MSN precursor cells. In addition, conditional overexpression of Six3 promoted the differentiation of precursor cells in the LGE. We measured an increase of apoptosis in the postnatal striatum of conditional Six3-knockout mice. This suggests that, in the absence of Six3, abnormally differentiated D2 MSNs are eliminated by programmed cell death. These results further identify Six3 as an important regulatory element during D2 MSN differentiation.
Keywords: Drd2; LGE; Medium spiny neuron; Six3; striatum.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Dlx1/2-dependent expression of Meis2 promotes neuronal fate determination in the mammalian striatum.Development. 2022 Feb 15;149(4):dev200035. doi: 10.1242/dev.200035. Epub 2022 Feb 23. Development. 2022. PMID: 35156680 Free PMC article.
-
SP8 and SP9 coordinately promote D2-type medium spiny neuron production by activating Six3 expression.Development. 2018 Jul 23;145(14):dev165456. doi: 10.1242/dev.165456. Development. 2018. PMID: 29967281 Free PMC article.
-
Zfhx3 is required for the differentiation of late born D1-type medium spiny neurons.Exp Neurol. 2019 Dec;322:113055. doi: 10.1016/j.expneurol.2019.113055. Epub 2019 Sep 3. Exp Neurol. 2019. PMID: 31491374
-
D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons.Trends Neurosci. 2007 May;30(5):228-35. doi: 10.1016/j.tins.2007.03.008. Epub 2007 Apr 3. Trends Neurosci. 2007. PMID: 17408758 Review.
-
A role for TGFβ signalling in medium spiny neuron differentiation of human pluripotent stem cells.Neuronal Signal. 2020 May 6;4(2):NS20200004. doi: 10.1042/NS20200004. eCollection 2020 Jun. Neuronal Signal. 2020. PMID: 32714602 Free PMC article. Review.
Cited by
-
Spatial enhancer activation influences inhibitory neuron identity during mouse embryonic development.Nat Neurosci. 2024 May;27(5):862-872. doi: 10.1038/s41593-024-01611-9. Epub 2024 Mar 25. Nat Neurosci. 2024. PMID: 38528203 Free PMC article.
-
The transcriptomic and spatial organization of telencephalic GABAergic neuronal types.bioRxiv [Preprint]. 2024 Jun 18:2024.06.18.599583. doi: 10.1101/2024.06.18.599583. bioRxiv. 2024. PMID: 38948843 Free PMC article. Preprint.
-
The transcription factor Zfp503 promotes the D1 MSN identity and represses the D2 MSN identity.Front Cell Dev Biol. 2022 Aug 23;10:948331. doi: 10.3389/fcell.2022.948331. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36081908 Free PMC article.
-
Chd5 Regulates the Transcription Factor Six3 to Promote Neuronal Differentiation.Stem Cells. 2023 Mar 17;41(3):242-251. doi: 10.1093/stmcls/sxad002. Stem Cells. 2023. PMID: 36636025 Free PMC article.
-
Dlx1/2-dependent expression of Meis2 promotes neuronal fate determination in the mammalian striatum.Development. 2022 Feb 15;149(4):dev200035. doi: 10.1242/dev.200035. Epub 2022 Feb 23. Development. 2022. PMID: 35156680 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

