Immunity onset alters plant chromatin and utilizes EDA16 to regulate oxidative homeostasis
- PMID: 34015058
- PMCID: PMC8171942
- DOI: 10.1371/journal.ppat.1009572
Immunity onset alters plant chromatin and utilizes EDA16 to regulate oxidative homeostasis
Abstract
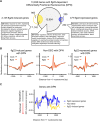
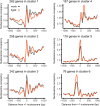
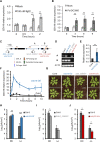
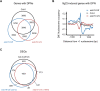
Perception of microbes by plants leads to dynamic reprogramming of the transcriptome, which is essential for plant health. The appropriate amplitude of this transcriptional response can be regulated at multiple levels, including chromatin. However, the mechanisms underlying the interplay between chromatin remodeling and transcription dynamics upon activation of plant immunity remain poorly understood. Here, we present evidence that activation of plant immunity by bacteria leads to nucleosome repositioning, which correlates with altered transcription. Nucleosome remodeling follows distinct patterns of nucleosome repositioning at different loci. Using a reverse genetic screen, we identify multiple chromatin remodeling ATPases with previously undescribed roles in immunity, including EMBRYO SAC DEVELOPMENT ARREST 16, EDA16. Functional characterization of the immune-inducible chromatin remodeling ATPase EDA16 revealed a mechanism to negatively regulate immunity activation and limit changes in redox homeostasis. Our transcriptomic data combined with MNase-seq data for EDA16 functional knock-out and over-expressor mutants show that EDA16 selectively regulates a defined subset of genes involved in redox signaling through nucleosome repositioning. Thus, collectively, chromatin remodeling ATPases fine-tune immune responses and provide a previously uncharacterized mechanism of immune regulation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Lewis LA, Polanski K, de Torres-Zabala M, Jayaraman S, Bowden L, Moore J, et al.. Transcriptional Dynamics Driving MAMP-Triggered Immunity and Pathogen Effector-Mediated Immunosuppression in Arabidopsis Leaves Following Infection with Pseudomonas syringae pv tomato DC3000. Plant Cell. 2015;27: 3038–3064. 10.1105/tpc.15.00471 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

