Elevated expression of LPCAT1 predicts a poor prognosis and is correlated with the tumour microenvironment in endometrial cancer
- PMID: 34016103
- PMCID: PMC8139085
- DOI: 10.1186/s12935-021-01965-1
Elevated expression of LPCAT1 predicts a poor prognosis and is correlated with the tumour microenvironment in endometrial cancer
Abstract
Background: Endometrial cancer (EC) is one of the three malignant reproductive tumours that threaten women's lives and health. Glycerophospholipids (GPLs) are important bioactive lipids involved in various physiological and pathological processes, including cancer. Immune infiltration of the tumour microenvironment (TME) is positively associated with the overall survival in EC. Exploring GPL-related factors associated with the TME in endometrial cancer can aid in the prognosis of patients and provide new therapeutic targets.
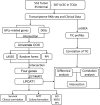
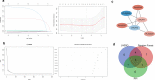
Methods: Differentially expressed GPL-related genes were identified from TCGA-UCEC datasets and the Molecular Signatures Database (MSigDB). Univariate Cox regression analysis was used to select GPL-related genes with prognostic value. The Random forest algorithm, LASSO algorithm and PPI network were used to identify critical genes. ESTIMATEScore was calculated to identify genes associated with the TME. Then, differentiation analysis and survival analysis of LPCAT1 were performed based on TCGA datasets. GSE17025 and immunohistochemistry (IHC) verified the results of the differentiation analysis. An MTT assay was then conducted to determine the proliferation of EC cells. GO and KEGG enrichment analyses were performed to explore the underlying mechanism of LPCAT1. In addition, we used the ssGSEA algorithm to explore the correlation between LPCAT1 and cancer immune infiltrates.
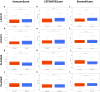
Results: Twenty-three differentially expressed GPL-related genes were identified, and eleven prognostic genes were selected by univariate Cox regression analysis. Four significant genes were identified by two different algorithms and the PPI network. Only LPCAT1 was significantly correlated with the tumour microenvironment. Then, we found that LPCAT1 was highly expressed in tumour samples compared with that in normal tissues, and lower survival rates were observed in the groups with high LPCAT1 expression. Silencing of LPCAT1 inhibited the proliferation of EC cells. Moreover, the expression of LPCAT1 was positively correlated with the histologic grades and types. The ROC curve indicated that LPCAT1 had good prognostic accuracy. Receptor ligand activity, pattern specification process, regionalization, anterior/posterior pattern specification and salivary secretion pathways were enriched as potential targets of LPCAT1. By using the ssGSEA algorithm, fifteen kinds of tumor-infiltrating cells (TICs) were found to be correlated with LPCAT1 expression.
Conclusion: These findings suggested that LPCAT1 may act as a valuable prognostic biomarker and be correlated with immune infiltrates in endometrial cancer, which may provide novel therapy options for and improved treatment of EC.
Keywords: Endometrial cancer; Glycerophospholipids; LPCAT1; Tumor microenvironment.
Conflict of interest statement
The authors declare no competing interests.
Figures
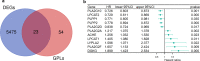


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

