Toward a Phage Cocktail for Tuberculosis: Susceptibility and Tuberculocidal Action of Mycobacteriophages against Diverse Mycobacterium tuberculosis Strains
- PMID: 34016711
- PMCID: PMC8263002
- DOI: 10.1128/mBio.00973-21
Toward a Phage Cocktail for Tuberculosis: Susceptibility and Tuberculocidal Action of Mycobacteriophages against Diverse Mycobacterium tuberculosis Strains
Abstract
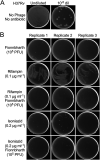
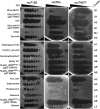
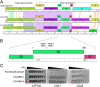
The global health burden of human tuberculosis (TB) and the widespread antibiotic resistance of its causative agent Mycobacterium tuberculosis warrant new strategies for TB control. The successful use of a bacteriophage cocktail to treat a Mycobacterium abscessus infection suggests that phages could play a role in tuberculosis therapy. To assemble a phage cocktail with optimal therapeutic potential for tuberculosis, we have explored mycobacteriophage diversity to identify phages that demonstrate tuberculocidal activity and determined the phage infection profiles for a diverse set of strains spanning the major lineages of human-adapted strains of the Mycobacterium tuberculosis complex. Using a combination of genome engineering and bacteriophage genetics, we have assembled a five-phage cocktail that minimizes the emergence of phage resistance and cross-resistance to multiple phages, and which efficiently kills the M. tuberculosis strains tested. Furthermore, these phages function without antagonizing antibiotic effectiveness, and infect both isoniazid-resistant and -sensitive strains.IMPORTANCE Tuberculosis kills 1.5 million people each year, and resistance to commonly used antibiotics contributes to treatment failures. The therapeutic potential of bacteriophages against Mycobacterium tuberculosis offers prospects for shortening antibiotic regimens, provides new tools for treating multiple drug-resistant (MDR)-TB and extensively drug-resistant (XDR)-TB infections, and protects newly developed antibiotics against rapidly emerging resistance to them. Identifying a suitable suite of phages active against diverse M. tuberculosis isolates circumvents many of the barriers to initiating clinical evaluation of phages as part of the arsenal of antituberculosis therapeutics.
Keywords: Mycobacterium tuberculosis; bacteriophage therapy; bacteriophages; tuberculosis.
Copyright © 2021 Guerrero-Bustamante et al.
Figures






References
-
- Centers for Disease Control. 2020. World TB Day 2020. https://www.cdc.gov/tb/worldtbday/history.htm.
-
- Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, ZiaZarifi AH, Hoffner SE. 2009. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in Iran. Chest 136:420–425. doi:10.1378/chest.08-2427. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

