Druggable epigenetic suppression of interferon-induced chemokine expression linked to MYCN amplification in neuroblastoma
- PMID: 34016720
- PMCID: PMC8141444
- DOI: 10.1136/jitc-2020-001335
Druggable epigenetic suppression of interferon-induced chemokine expression linked to MYCN amplification in neuroblastoma
Abstract
Background: Amplification of the MYCN oncogene is a molecular hallmark of aggressive neuroblastoma (NB), a childhood cancer of the sympathetic nervous system. There is evidence that MYCN promotes a non-inflamed and T-cell infiltration-poor ('cold') tumor microenvironment (TME) by suppressing interferon signaling. This may explain, at least in part, why patients with NB seem to have little benefit from single-agent immune checkpoint blockade (ICB) therapy. Targeting MYCN or its effectors could be a strategy to convert a cold TME into a 'hot' (inflamed) TME and improve the efficacy of ICB therapy.
Methods: NB transcriptome analyses were used to identify epigenetic drivers of a T-cell infiltration-poor TME. Biological and molecular responses of NB cells to epigenetic drugs and interferon (IFN)-γ exposure were assessed by proliferation assays, immunoblotting, ELISA, qRT-PCR, RNA-seq and ChIP-qPCR as well as co-culture assays with T cells.
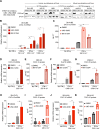
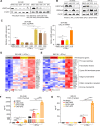
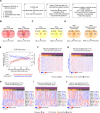
Results: We identified H3K9 euchromatic histone-lysine methyltransferases EHMT2 and EHMT1, also known as G9a and GLP, as epigenetic effectors of the MYCN-driven malignant phenotype and repressors of IFN-γ transcriptional responses in NB cells. EHMT inhibitors enhanced IFN-γ-induced expression of the Th1-type chemokines CXCL9 and CXCL10, key factors of T-cell recruitment into the TME. In MYCN-amplified NB cells, co-inhibition of EZH2 (enhancer of zeste homologue 2), a H3K27 histone methyltransferase cooperating with EHMTs, was needed for strong transcriptional responses to IFN-γ, in line with histone mark changes at CXCL9 and CXCL10 chemokine gene loci. EHMT and EZH2 inhibitor response gene signatures from NB cells were established as surrogate measures and revealed high EHMT and EZH2 activity in MYCN-amplified high-risk NBs with a cold immune phenotype.
Conclusion: Our results delineate a strategy for targeted epigenetic immunomodulation of high-risk NBs, whereby EHMT inhibitors alone or in combination with EZH2 inhibitors (in particular, MYCN-amplified NBs) could promote a T-cell-infiltrated TME via enhanced Th1-type chemokine expression.
Keywords: immunomodulation; immunotherapy; inflammation; neuroblastoma; tumor microenvironment.
© Author(s) (or their employer(s)) 2021. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




Similar articles
-
Epigenetic Dysregulation in MYCN-Amplified Neuroblastoma.Int J Mol Sci. 2023 Dec 3;24(23):17085. doi: 10.3390/ijms242317085. Int J Mol Sci. 2023. PMID: 38069407 Free PMC article. Review.
-
Histone-lysine methyltransferase EHMT2 is involved in proliferation, apoptosis, cell invasion, and DNA methylation of human neuroblastoma cells.Anticancer Drugs. 2013 Jun;24(5):484-93. doi: 10.1097/CAD.0b013e32835ffdbb. Anticancer Drugs. 2013. PMID: 23466651 Free PMC article.
-
GD2 redirected CAR T and activated NK-cell-mediated secretion of IFNγ overcomes MYCN-dependent IDO1 inhibition, contributing to neuroblastoma cell immune escape.J Immunother Cancer. 2021 Mar;9(3):e001502. doi: 10.1136/jitc-2020-001502. J Immunother Cancer. 2021. PMID: 33737337 Free PMC article.
-
Transcript signatures that predict outcome and identify targetable pathways in MYCN-amplified neuroblastoma.Mol Oncol. 2016 Nov;10(9):1461-1472. doi: 10.1016/j.molonc.2016.07.012. Epub 2016 Aug 18. Mol Oncol. 2016. PMID: 27599694 Free PMC article.
-
T cells in the microenvironment of solid pediatric tumors: the case of neuroblastoma.Front Immunol. 2025 Feb 28;16:1544137. doi: 10.3389/fimmu.2025.1544137. eCollection 2025. Front Immunol. 2025. PMID: 40092980 Free PMC article. Review.
Cited by
-
Epigenetic Dysregulation in MYCN-Amplified Neuroblastoma.Int J Mol Sci. 2023 Dec 3;24(23):17085. doi: 10.3390/ijms242317085. Int J Mol Sci. 2023. PMID: 38069407 Free PMC article. Review.
-
Progress Toward Epigenetic Targeted Therapies for Childhood Cancer.Cancers (Basel). 2024 Dec 12;16(24):4149. doi: 10.3390/cancers16244149. Cancers (Basel). 2024. PMID: 39766049 Free PMC article. Review.
-
Lysine methylation modifications in tumor immunomodulation and immunotherapy: regulatory mechanisms and perspectives.Biomark Res. 2024 Jul 30;12(1):74. doi: 10.1186/s40364-024-00621-w. Biomark Res. 2024. PMID: 39080807 Free PMC article. Review.
-
Epigenetic modulation of neuroblastoma enhances T cell and NK cell immunogenicity by inducing a tumor-cell lineage switch.J Immunother Cancer. 2022 Dec;10(12):e005002. doi: 10.1136/jitc-2022-005002. J Immunother Cancer. 2022. PMID: 36521927 Free PMC article.
-
Targeted immune activation in pediatric solid tumors: opportunities to complement local control approaches.Front Immunol. 2023 Jun 22;14:1202169. doi: 10.3389/fimmu.2023.1202169. eCollection 2023. Front Immunol. 2023. PMID: 37426669 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
