Epidermal chloroplasts are defense-related motile organelles equipped with plant immune components
- PMID: 34016974
- PMCID: PMC8137707
- DOI: 10.1038/s41467-021-22977-5
Epidermal chloroplasts are defense-related motile organelles equipped with plant immune components
Abstract
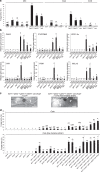
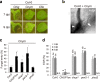
In addition to conspicuous large mesophyll chloroplasts, where most photosynthesis occurs, small epidermal chloroplasts have also been observed in plant leaves. However, the functional significance of this small organelle remains unclear. Here, we present evidence that Arabidopsis epidermal chloroplasts control the entry of fungal pathogens. In entry trials, specialized fungal cells called appressoria triggered dynamic movement of epidermal chloroplasts. This movement is controlled by common regulators of mesophyll chloroplast photorelocation movement, designated as the epidermal chloroplast response (ECR). The ECR occurs when the PEN2 myrosinase-related higher-layer antifungal system becomes ineffective, and blockage of the distinct steps of the ECR commonly decreases preinvasive nonhost resistance against fungi. Furthermore, immune components were preferentially localized to epidermal chloroplasts, contributing to antifungal nonhost resistance in the pen2 background. Our findings reveal that atypical small chloroplasts act as defense-related motile organelles by specifically positioning immune components in the plant epidermis, which is the first site of contact between the plant and pathogens. Thus, this work deepens our understanding of the functions of epidermal chloroplasts.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Kubo, Y. & Furusawa, I. The Fungal Spore and Disease Initiation in Plants and Animals (eds Cole, G.T. & Hoch, H.C.) (Plenum Publishing, 1991).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

